5MKG
 
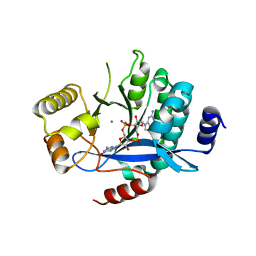 | | PA3825-EAL Ca-CdG Structure | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | 著者 | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | 登録日 | 2016-12-04 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
4DQH
 
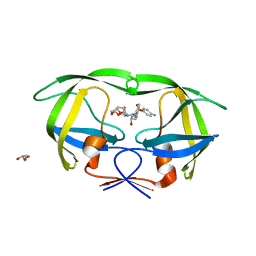 | | Crystal Structure of (R14C/E65C) HIV-1 Protease in complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Wild-type HIV-1 protease dimer | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DQB
 
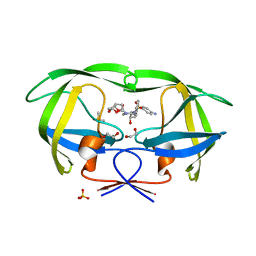 | | Crystal Structure of wild-type HIV-1 Protease in Complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DQC
 
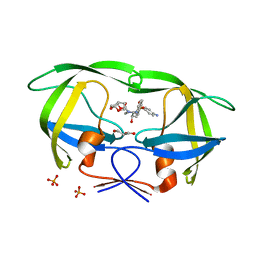 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Aspartyl protease, GLYCEROL, ... | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Chen, S.J, Ye, F. | | 登録日 | 2016-08-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GUV
 
 | |
4K24
 
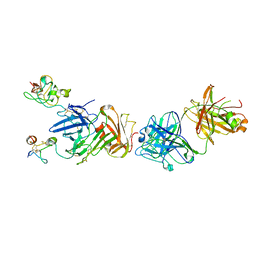 | | Structure of anti-uPAR Fab ATN-658 in complex with uPAR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | 著者 | Huang, M.D, Xu, X, Yuan, C. | | 登録日 | 2013-04-08 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
4QGE
 
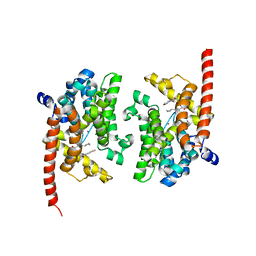 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | 分子名称: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | 著者 | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | 登録日 | 2014-05-22 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
4K23
 
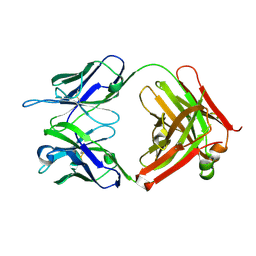 | | Structure of anti-uPAR Fab ATN-658 | | 分子名称: | anti-uPAR antibody, heavy chain, light chain | | 著者 | Yuan, C, Huang, M, Chen, L. | | 登録日 | 2013-04-08 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
5CFF
 
 | |
7T63
 
 | |
4Y9P
 
 | | PA3825-EAL Ca-CdG Structure | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, PA3825-EAL | | 著者 | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | 登録日 | 2015-02-17 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
4Y9M
 
 | | PA3825-EAL Metal-Free-Apo Structure | | 分子名称: | PA3825-EAL, PHOSPHATE ION | | 著者 | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | 登録日 | 2015-02-17 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
4GH6
 
 | | Crystal structure of the PDE9A catalytic domain in complex with inhibitor 28 | | 分子名称: | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, N-(4-methoxyphenyl)-N~2~-[1-(2-methylphenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]-L-alaninamide, ... | | 著者 | Hou, J, Ke, H. | | 登録日 | 2012-08-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Based Discovery of Highly Selective Phosphodiesterase-9A Inhibitors and Implications for Inhibitor Design.
J.Med.Chem., 55, 2012
|
|
6B20
 
 | | Crystal structure of a complex between G protein beta gamma dimer and an inhibitory Nanobody regulator | | 分子名称: | CHLORIDE ION, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | 著者 | Gulati, S, Kiser, P.D, Palczewski, K. | | 登録日 | 2017-09-19 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor.
Nat Commun, 9, 2018
|
|
3P57
 
 | | Crystal structure of the p300 TAZ2 domain bound to MEF2 on DNA | | 分子名称: | DNA (5'-D(*A*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), Histone acetyltransferase p300, ... | | 著者 | He, J, Ye, J, Riquelme, C, Liu, J.O. | | 登録日 | 2010-10-08 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1921 Å) | | 主引用文献 | Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly.
Nucleic Acids Res., 39, 2011
|
|
8WVD
 
 | | Crystal structure of Glycosyltransferase in complex with UD1 | | 分子名称: | Glycosyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2023-10-23 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Enzymatic Synthesis of Novel Terpenoid Glycoside Derivatives Decorated with N -Acetylglucosamine Catalyzed by UGT74AC1.
J.Agric.Food Chem., 72, 2024
|
|
3TW8
 
 | |
8WX7
 
 | | Crystal structure of SHP2 in complex with JAB-3186 | | 分子名称: | (5~{S})-1'-[6-azanyl-5-(2-azanyl-3-chloranyl-pyridin-4-yl)sulfanyl-pyrazin-2-yl]spiro[5,7-dihydrocyclopenta[b]pyridine-6,4'-piperidine]-5-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Ma, C, Gao, P, Kang, D, Han, H, Sun, X, Zhang, W, Qian, D, Wang, Y, Long, W. | | 登録日 | 2023-10-27 | | 公開日 | 2024-08-14 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Discovery of JAB-3312, a Potent SHP2 Allosteric Inhibitor for Cancer Treatment.
J.Med.Chem., 67, 2024
|
|
4DQF
 
 | | Crystal Structure of (G16A/L38A) HIV-1 Protease in Complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
7W2Z
 
 | | Cryo-EM structure of the ghrelin-bound human ghrelin receptor-Go complex | | 分子名称: | Appetite-regulating hormone, CHOLESTEROL, Growth hormone secretagogue receptor type 1, ... | | 著者 | Qin, J, Ming, Q, Ji, S, Mao, C, Shen, D, Zhang, Y. | | 登録日 | 2021-11-24 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DQG
 
 | |
6JUE
 
 | | The complex of PDZ and PBM | | 分子名称: | Partitioning defective 3 homolog, SULFATE ION, THR-ILE-ILE-THR-LEU | | 著者 | Liu, Z. | | 登録日 | 2019-04-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | Par complex cluster formation mediated by phase separation.
Nat Commun, 11, 2020
|
|
6L8Z
 
 | | Crystal structure of ugt transferase mutant in complex with UPG | | 分子名称: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
