3C33
 
 | |
3C36
 
 | |
3C34
 
 | |
5OSB
 
 | | GLIC-GABAAR alpha1 chimera crystallized in complex with THDOC at pH4.5 | | 分子名称: | ACETATE ION, CHLORIDE ION, Proton-gated ion channel,Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | 著者 | Laverty, D.C, Gold, M.G, Smart, T.G. | | 登録日 | 2017-08-17 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OSC
 
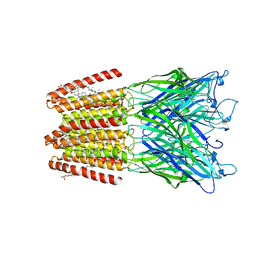 | | GLIC-GABAAR alpha1 chimera crystallized in complex with pregnenolone sulfate at pH 4.5 | | 分子名称: | ACETATE ION, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Laverty, D.C, Gold, M.G, Smart, T.G. | | 登録日 | 2017-08-17 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OSA
 
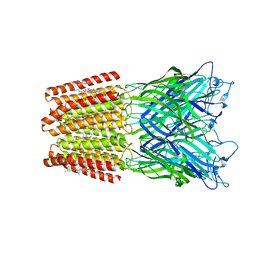 | | GLIC-GABAAR alpha1 chimera crystallized at pH4.6 | | 分子名称: | ACETATE ION, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Laverty, D.C, Gold, M.G, Smart, T.G. | | 登録日 | 2017-08-17 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.75000238 Å) | | 主引用文献 | Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6Y7V
 
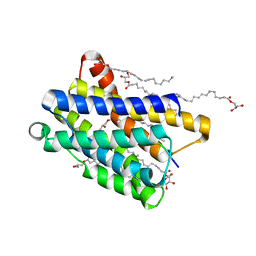 | |
6Z4A
 
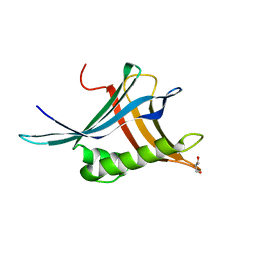 | |
6Z3T
 
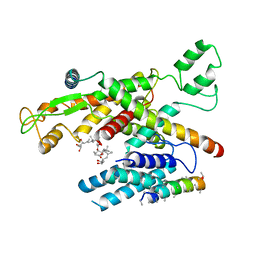 | | Structure of canine Sec61 inhibited by mycolactone | | 分子名称: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | 著者 | Gerard, S.F, Higgins, M.K. | | 登録日 | 2020-05-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.69 Å) | | 主引用文献 | Structure of the Inhibited State of the Sec Translocon.
Mol.Cell, 79, 2020
|
|
9BIT
 
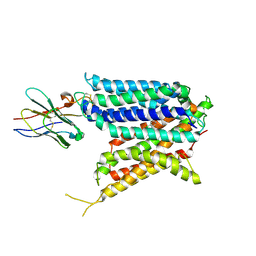 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 1 | | 分子名称: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | 著者 | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | 登録日 | 2024-04-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIU
 
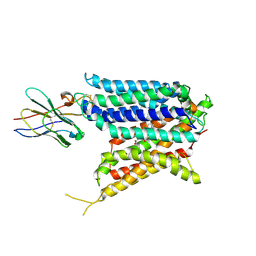 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 2 | | 分子名称: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | 著者 | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | 登録日 | 2024-04-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIS
 
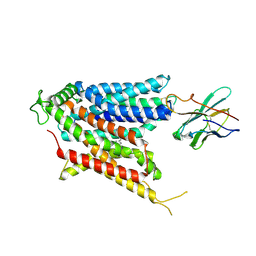 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Solute carrier family 15 member 2, nanobody | | 著者 | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | 登録日 | 2024-04-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIR
 
 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cefadroxil | | 分子名称: | Cefadroxil, Solute carrier family 15 member 2, nanobody | | 著者 | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | 登録日 | 2024-04-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
8I41
 
 | |
8I42
 
 | |
8I48
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-01-18 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8I47
 
 | |
7ZDD
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K10ac histone peptide. | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM33, Histone H3.X, ... | | 著者 | Caria, S, Duclos, S, Crespillo, S, Errey, J, Barker, J.J. | | 登録日 | 2022-03-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.625 Å) | | 主引用文献 | Identification of Histone Peptide Binding Specificity and Small-Molecule Ligands for the TRIM33 alpha and TRIM33 beta Bromodomains.
Acs Chem.Biol., 17, 2022
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
5FTH
 
 | |
8APY
 
 | | Crystal structure of the H12A variant of the KDEL receptor bound to sybody | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ER lumen protein-retaining receptor 2, Synthetic nanobody | | 著者 | Parker, J.L, Smith, K, Newstead, S. | | 登録日 | 2022-08-10 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Molecular basis for pH sensing in the KDEL trafficking receptor.
Structure, 32, 2024
|
|
6FT3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
5FTI
 
 | |
7SAB
 
 | | Phencyclidine-bound GluN1a-GluN2B NMDA receptors | | 分子名称: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chou, T.-H, Furukawa, H. | | 登録日 | 2021-09-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
