7XOC
 
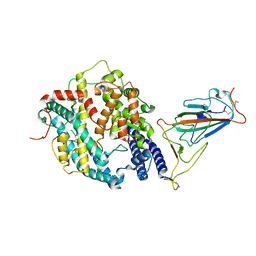 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | 登録日 | 2022-05-01 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOB
 
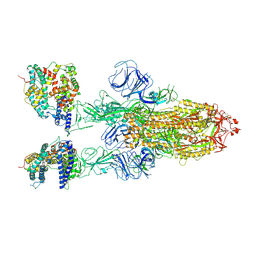 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | 登録日 | 2022-05-01 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | 著者 | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-02-22 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
7SVT
 
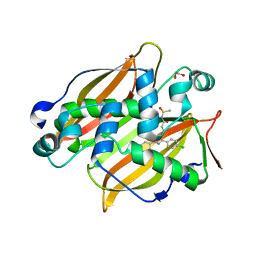 | | Mycobacterium tuberculosis 3-hydroxyl-ACP dehydratase HadAB in complex with 1,3-diarylpyrazolyl-acylsulfonamide inhibitor | | 分子名称: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 1,2-ETHANEDIOL, 3-[1-(4-bromophenyl)-3-(4-chlorophenyl)-1H-pyrazol-4-yl]-N-(methanesulfonyl)propanamide, ... | | 著者 | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2021-11-19 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 1,3-Diarylpyrazolyl-acylsulfonamides Target HadAB/BC Complex in Mycobacterium tuberculosis .
Acs Infect Dis., 8, 2022
|
|
7C4U
 
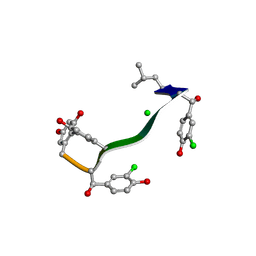 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | 分子名称: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | 著者 | Fan, Q, Zhou, H, Li, X, Wang, J. | | 登録日 | 2020-05-18 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | 主引用文献 | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | 分子名称: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | 著者 | Fan, Q, Zhou, H, Li, X, Wang, J. | | 登録日 | 2020-05-18 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | 主引用文献 | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8GNJ
 
 | |
8GNI
 
 | |
8GQ5
 
 | |
8HS2
 
 | | Orphan GPR20 in complex with Fab046 | | 分子名称: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
7UEK
 
 | |
8HS3
 
 | | Gi bound orphan GPR20 in ligand-free state | | 分子名称: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-19 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8H85
 
 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | 分子名称: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | 著者 | Gong, W.M, Hu, X.Y. | | 登録日 | 2022-10-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | 分子名称: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | 著者 | Gong, W.M, Hu, X.Y. | | 登録日 | 2022-10-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | 分子名称: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | 著者 | Gong, W.M, Hu, X.Y. | | 登録日 | 2022-10-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | 著者 | Gong, W.M, Hu, X.Y. | | 登録日 | 2022-10-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7V
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | 著者 | Gong, W.M, Hu, X.Y. | | 登録日 | 2022-10-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6LXI
 
 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | 分子名称: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | 著者 | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXJ
 
 | | Crystal structure of human Z2B3 Fab in complex with influenza virus neuraminidase from A/Anhui/1/2013 (H7N9) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Heavy chain of Z2B3 Fab, ... | | 著者 | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.903 Å) | | 主引用文献 | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | 分子名称: | Putative periplasmic protein | | 著者 | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
6LXK
 
 | | Crystal structure of Z2B3 D102R Fab in complex with influenza virus neuraminidase from A/Serbia/NS-601/2014 (H1N1) | | 分子名称: | CALCIUM ION, Heavy chain of Z2B3-D102R Fab, Light chain of Z2B3-D102R Fab, ... | | 著者 | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.608 Å) | | 主引用文献 | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
7W27
 
 | | Crystal structure of BEND3-BEN4-DNA complex | | 分子名称: | BEN domain-containing protein 3, DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*CP*GP*TP*GP*GP*GP*TP*C)-3') | | 著者 | Zheng, L, Ren, A. | | 登録日 | 2021-11-22 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Genes Dev., 36, 2022
|
|
5YGU
 
 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | 分子名称: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | 登録日 | 2017-09-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
2L93
 
 | |
