3RHG
 
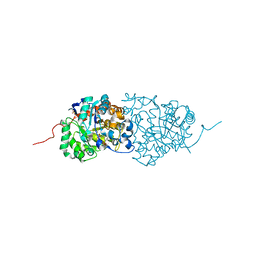 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | 分子名称: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | 著者 | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-04-11 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3T61
 
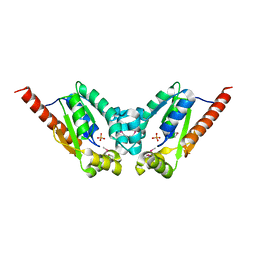 | | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021 | | 分子名称: | Gluconokinase, PHOSPHATE ION | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-07-28 | | 公開日 | 2011-08-17 | | 最終更新日 | 2012-03-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021
To be Published
|
|
3SY5
 
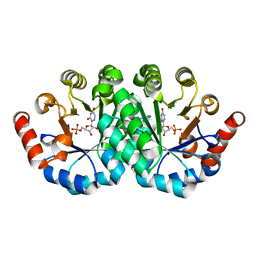 | | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor 6azaUMP | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Desai, B, Iiams, V, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-07-15 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.321 Å) | | 主引用文献 | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor 6azaUMP
To be Published
|
|
3T8Q
 
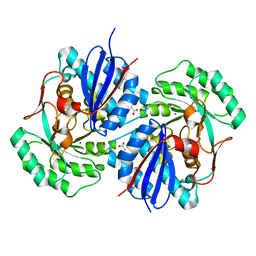 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica | | 分子名称: | MAGNESIUM ION, MALONATE ION, Mandelate racemase/muconate lactonizing enzyme family protein | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-08-01 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica
To be Published
|
|
3RDM
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin/PEG | | 分子名称: | BIOTIN, PENTAETHYLENE GLYCOL, Streptavidin | | 著者 | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | 登録日 | 2011-04-01 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDU
 
 | | Crystal structure of R7-2 streptavidin complexed with PEG | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | 著者 | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | 登録日 | 2011-04-01 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RN6
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and isoguanine | | 分子名称: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 6-amino-3,7-dihydro-2H-purin-2-one, Cytosine deaminase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | 登録日 | 2011-04-22 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.255 Å) | | 主引用文献 | Rescue of the orphan enzyme isoguanine deaminase.
Biochemistry, 50, 2011
|
|
3RHA
 
 | |
3QU7
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with calcium and phosphate | | 分子名称: | ACETATE ION, CALCIUM ION, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU5
 
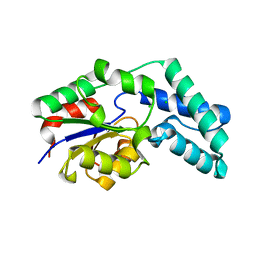 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp11asn mutant | | 分子名称: | CHLORIDE ION, INORGANIC PYROPHOSPHATASE | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU2
 
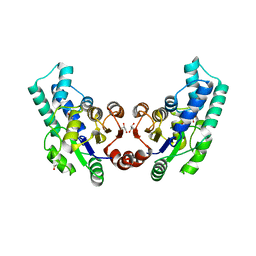 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, a closed cap conformation | | 分子名称: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUT
 
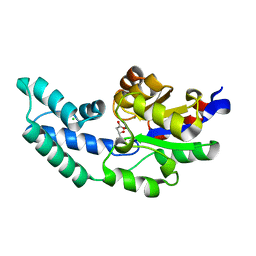 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant, an open cap conformation | | 分子名称: | CHLORIDE ION, D-MALATE, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-24 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUC
 
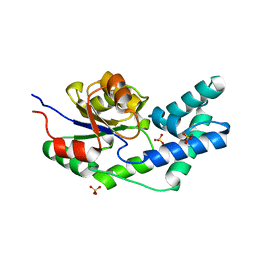 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asn mutant complexed with sulfate | | 分子名称: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QYP
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asn mutant complexed with calcium and phosphate | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-03-03 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3R09
 
 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | 分子名称: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2011-03-07 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
3R0N
 
 | | Crystal Structure of the Immunoglobulin variable domain of Nectin-2 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Poliovirus receptor-related protein 2 | | 著者 | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2011-03-08 | | 公開日 | 2011-04-27 | | 最終更新日 | 2012-11-07 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell-cell adhesion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R0U
 
 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Tartrate and Mg complex | | 分子名称: | D(-)-TARTARIC ACID, Enzyme of enolase superfamily, GLYCEROL, ... | | 著者 | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-03-09 | | 公開日 | 2011-04-06 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | 分子名称: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | 著者 | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | 登録日 | 2011-04-01 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.404 Å) | | 主引用文献 | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | 著者 | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | 登録日 | 2011-04-02 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3TFO
 
 | | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti | | 分子名称: | HEXANE-1,6-DIOL, SULFATE ION, putative 3-oxoacyl-(acyl-carrier-protein) reductase | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-08-16 | | 公開日 | 2011-08-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti
To be Published
|
|
3T66
 
 | | Crystal structure of Nickel ABC transporter from Bacillus halodurans | | 分子名称: | CALCIUM ION, Nickel ABC transporter (Nickel-binding protein) | | 著者 | Agarwal, R, Bonanno, J.B, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-07-28 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Nickel ABC transporter from Bacillus halodurans
To be Published
|
|
3R6A
 
 | |
3RLU
 
 | | Crystal structure of the mutant K82A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-04-20 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3RMT
 
 | | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125 | | 分子名称: | 3-phosphoshikimate 1-carboxyvinyltransferase 1, SULFATE ION | | 著者 | Malashkevich, V.N, Toro, R, Seidel, R, Ramagopal, U, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-04-21 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125
To be Published
|
|
3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | 分子名称: | Leukocyte-associated immunoglobulin-like receptor 1 | | 著者 | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-04-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
