1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-05-19 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
2FZ1
 
 | |
1M6Z
 
 | | Crystal structure of reduced recombinant cytochrome c4 from Pseudomonas stutzeri | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome c4, GLYCEROL, ... | | 著者 | Noergaard, A, Harris, P, Larsen, S, Christensen, H.E.M. | | 登録日 | 2002-07-18 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural comparison of recombinant Pseudomonas stutzeri cytochrome c4 in two oxidation states
To be Published
|
|
2D22
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2JRA
 
 | | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris. Northeast Structural Genomics Target RpT6 | | 分子名称: | Protein RPA2121 | | 著者 | Wu, B, Yee, A, Lemak, A, Cort, J, Bansal, S, Semest, A, Guido, V, Kennedy, M.A, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-21 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris.
To be Published
|
|
1DAR
 
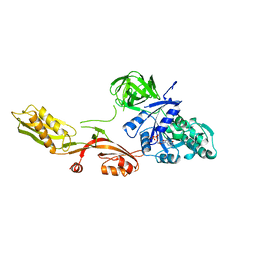 | | ELONGATION FACTOR G IN COMPLEX WITH GDP | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Al-Karadaghi, S, Aevarsson, A, Garber, M, Zheltonosova, J, Liljas, A. | | 登録日 | 1996-02-15 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange.
Structure, 4, 1996
|
|
2D1Z
 
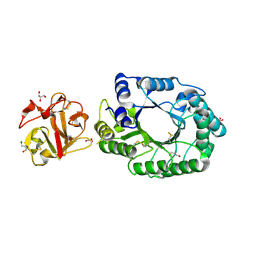 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
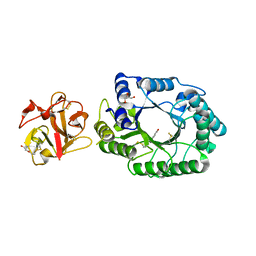 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
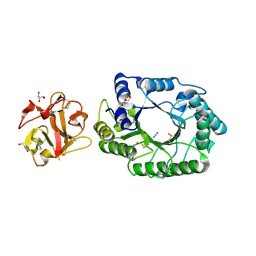 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2FWV
 
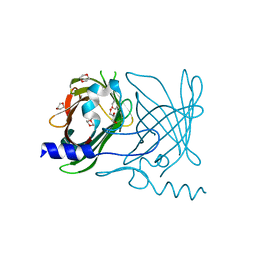 | | Crystal Structure of Rv0813 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, hypothetical protein MtubF_01000852 | | 著者 | Shepard, W, Haouz, A, Grana, M, Buschiazzo, A, Betton, J.M, Cole, S.T, Alzari, P.M, Structural Proteomics in Europe (SPINE) | | 登録日 | 2006-02-03 | | 公開日 | 2006-08-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of Rv0813c from Mycobacterium tuberculosis Reveals a New Family of Fatty Acid-Binding Protein-Like Proteins in Bacteria
J.Bacteriol., 189, 2007
|
|
2FSN
 
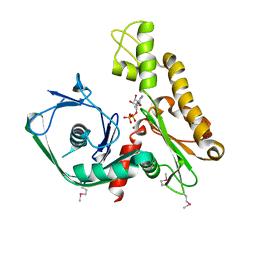 | | Crystal structure of Ta0583, an archaeal actin homolog, complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein Ta0583 | | 著者 | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | 登録日 | 2006-01-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2JCH
 
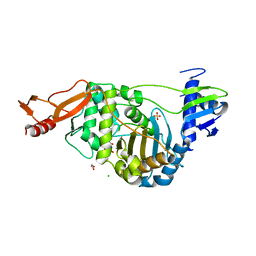 | | Structural and mechanistic basis of penicillin binding protein inhibition by lactivicins | | 分子名称: | (2E)-2-({(2S)-2-CARBOXY-2-[(PHENOXYACETYL)AMINO]ETHOXY}IMINO)PENTANEDIOIC ACID, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | 登録日 | 2006-12-23 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
2JIS
 
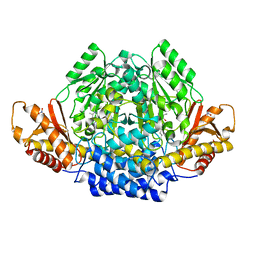 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | 分子名称: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | 登録日 | 2007-06-30 | | 公開日 | 2007-08-28 | | 最終更新日 | 2015-04-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
4JDW
 
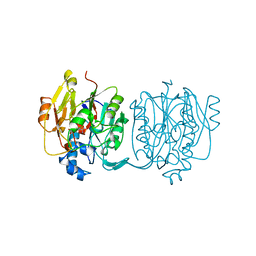 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | 分子名称: | ARGININE, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | 著者 | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | 登録日 | 1997-01-24 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1EL3
 
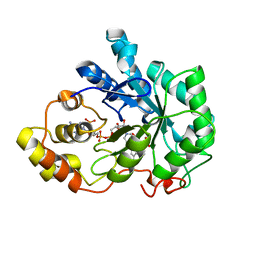 | | HUMAN ALDOSE REDUCTASE COMPLEXED WITH IDD384 INHIBITOR | | 分子名称: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,6-DIMETHYL-4-(2-O-TOLYL-ACETYLAMINO)-BENZENESULFONYL]-GLYCINE | | 著者 | Podjarny, A. | | 登録日 | 2000-03-13 | | 公開日 | 2000-05-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structure of human aldose reductase bound to the inhibitor IDD384.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EMS
 
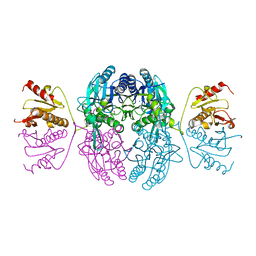 | | CRYSTAL STRUCTURE OF THE C. ELEGANS NITFHIT PROTEIN | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHYL MERCURY ION, NIT-FRAGILE HISTIDINE TRIAD FUSION PROTEIN, ... | | 著者 | Pace, H.C, Hodawadekar, S.C, Draganescu, A, Huang, J, Bieganowski, P, Pekarsky, Y, Croce, C.M, Brenner, C. | | 登録日 | 2000-03-17 | | 公開日 | 2000-07-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
Curr.Biol., 10, 2000
|
|
1EOS
 
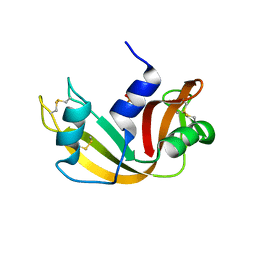 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A COMPLEXED WITH URIDYLYL(2',5')GUANOSINE (PRODUCTIVE BINDING) | | 分子名称: | RIBONUCLEASE PANCREATIC, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | 著者 | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | 登録日 | 2000-03-24 | | 公開日 | 2000-08-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Productive and nonproductive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2',5')guanosine.
Protein Sci., 9, 2000
|
|
2LEZ
 
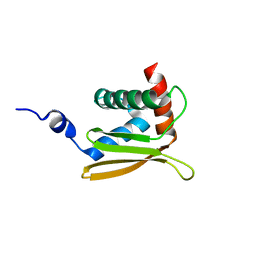 | | Solution NMR structure of N-terminal domain of Salmonella effector protein PipB2. Northeast structural genomics consortium (NESG) target stt318a | | 分子名称: | Secreted effector protein pipB2 | | 著者 | Lemak, A, Yee, A, Houliston, S, Garcia, M, Daniels, C, Savchenko, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-06-27 | | 公開日 | 2011-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of N-terminal domain of Salmonella effector protein PipB2
To be Published
|
|
2LBU
 
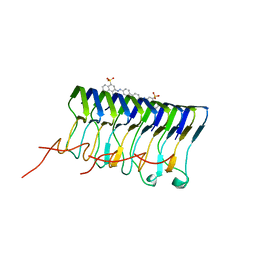 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | 分子名称: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | 著者 | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | 登録日 | 2011-04-07 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
2LGJ
 
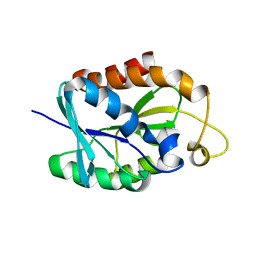 | | Solution structure of MsPTH | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | 登録日 | 2011-07-27 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
2LDC
 
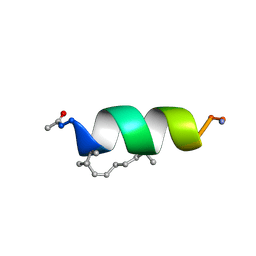 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | 分子名称: | Estrogen receptor-binding stapled peptide SP1 | | 著者 | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | 登録日 | 2011-05-20 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LST
 
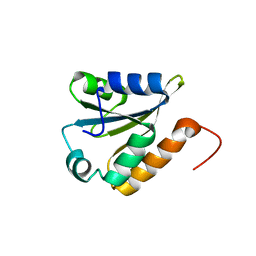 | | Solution structure of a thioredoxin from Thermus thermophilus | | 分子名称: | Thioredoxin | | 著者 | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-04 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a thioredoxin from Thermus thermophilus
To be Published
|
|
2LDD
 
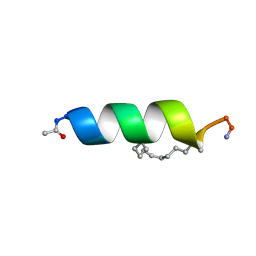 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | 分子名称: | Estrogen receptor-binding stapled peptide SP6 | | 著者 | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | 登録日 | 2011-05-21 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2L7R
 
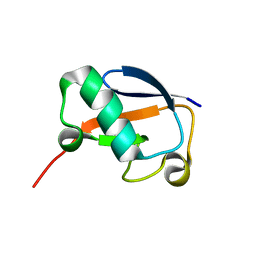 | | Solution NMR structure of N-terminal Ubiquitin-like domain of FUBI, a ribosomal protein S30 precursor from Homo sapiens. NorthEast Structural Genomics consortium (NESG) target HR6166 | | 分子名称: | Ubiquitin-like protein FUBI | | 著者 | Lemak, A, Yee, A, Houliston, S, Semesi, A, Doherty, R, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-20 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of N-terminal Ubiquitin-like domain of FUBI, a ribosomal protein S30
precursor from Homo sapiens. NorthEast Structural Genomics consortium (NESG) target HR6166
To be Published
|
|
6O3R
 
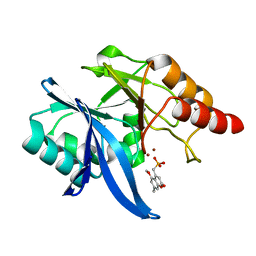 | | Crystal Structure of NDM-1 D199N with Compound 7 | | 分子名称: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5-methoxy-7-methyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2019-02-27 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
