5BN9
 
 | |
5BNH
 
 | | Crystal structure of the HLTF HIRAN domain with a ssDNA fragment | | 分子名称: | ACETATE ION, DNA (5'-D(*(GD)P*GP*TP*G)-3'), DNA (5'-D(*(TD)P*TP*G)-3'), ... | | 著者 | Neculai, D, Walker, J.R, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S. | | 登録日 | 2015-05-26 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Co-crystal structure of the HLTF HIRAN domain with a ssDNA fragment
To Be Published
|
|
5DAJ
 
 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | 分子名称: | NalD | | 著者 | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | 登録日 | 2015-08-20 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-04-12 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
1HKY
 
 | | Solution structure of a PAN module from Eimeria tenella | | 分子名称: | MICRONEME PROTEIN 5 PRECURSOR | | 著者 | Brown, P.J, Mulvey, D, Potts, J.R, Tomley, F.M, Campbell, I.D. | | 登録日 | 2002-10-03 | | 公開日 | 2002-10-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a Pan Module from the Apicomplexan Parasite Eimeria Tenella
J.Struct.Funct.Genom., 4, 2003
|
|
6HVF
 
 | | Kinase domain of cSrc in complex with compound 29B | | 分子名称: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, ~{N}-[3-[3-ethyl-6-[4-(4-methylpiperazin-1-yl)phenyl]-4-oxidanylidene-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]phenyl]prop-2-enamide | | 著者 | Keul, M, Mueller, M.P, Rauh, D. | | 登録日 | 2018-10-10 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibition of osimertinib-resistant epidermal growth factor receptor EGFR-T790M/C797S.
Chem Sci, 10, 2019
|
|
5D71
 
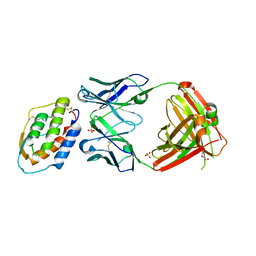 | | Crystal structure of MOR04302, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | 分子名称: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | 著者 | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | 登録日 | 2015-08-13 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
6HWT
 
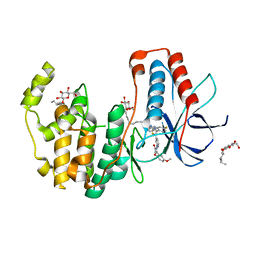 | | Crystal structure of p38alpha in complex with a reduced photoswitchable 2-Azothiazol-based Inhibitor (compound 31) | | 分子名称: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-(2-phenylhydrazinyl)-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Mueller, M.P, Rauh, D. | | 登録日 | 2018-10-15 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5D9B
 
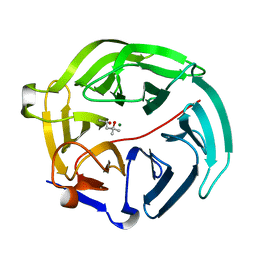 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | 著者 | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | 登録日 | 2015-08-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5CR1
 
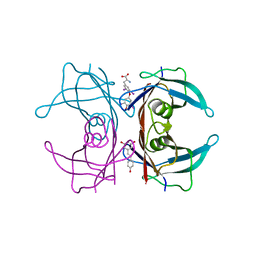 | | Crystal structure of TTR/resveratrol/T4 complex | | 分子名称: | 3,5,3',5'-TETRAIODO-L-THYRONINE, RESVERATROL, Transthyretin | | 著者 | Zanotti, G, Florio, P, Folli, C, Cianci, M, Del Rio, D, Berni, R. | | 登録日 | 2015-07-22 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.545 Å) | | 主引用文献 | Transthyretin Binding Heterogeneity and Anti-amyloidogenic Activity of Natural Polyphenols and Their Metabolites.
J.Biol.Chem., 290, 2015
|
|
6HZH
 
 | |
1HYB
 
 | | CRYSTAL STRUCTURE OF AN ACTIVE SITE MUTANT OF METHANOBACTERIUM THERMOAUTOTROPHICUM NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, SULFATE ION | | 著者 | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F. | | 登録日 | 2001-01-18 | | 公開日 | 2001-03-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
6HPI
 
 | |
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | 分子名称: | Urotensin-2 receptor | | 著者 | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | 登録日 | 2018-10-11 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
5DCN
 
 | | Crystal structure of LC3 in complex with TECPR2 LIR | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | 著者 | Stadel, D, Huber, J, Dotsch, V, Rogov, V.V, Behrends, C, Akutsu, M. | | 登録日 | 2015-08-24 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export.
Mol.Cell, 60, 2015
|
|
6I45
 
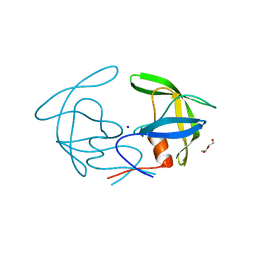 | | Crystal structure of I13V/I62V/V77I South African HIV-1 subtype C protease containing a D25A mutation | | 分子名称: | DI(HYDROXYETHYL)ETHER, Protease, SODIUM ION | | 著者 | Sherry, D, Pandian, R, Achilonu, I.A, Dirr, H.W, Sayed, Y. | | 登録日 | 2018-11-09 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Non-active site mutations in the HIV protease: Diminished drug binding affinity is achieved through modulating the hydrophobic sliding mechanism.
Int.J.Biol.Macromol., 217, 2022
|
|
6I1I
 
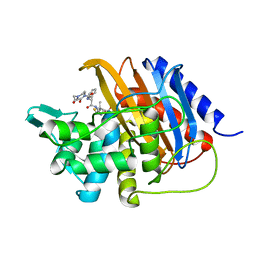 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | 分子名称: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-28 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
5CQ1
 
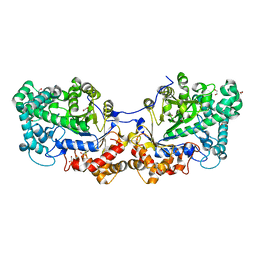 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | 分子名称: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | 著者 | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | 登録日 | 2015-07-21 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CMV
 
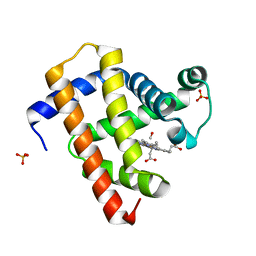 | | Ultrafast dynamics in myoglobin: dark-state, CO-ligated structure | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CND
 
 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6K42
 
 | | cryo-EM structure of alpha2BAR-Gi1 complex | | 分子名称: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | 登録日 | 2019-05-23 | | 公開日 | 2020-04-15 | | 最終更新日 | 2020-05-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
5CN5
 
 | | Ultrafast dynamics in myoglobin: 0 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5BWN
 
 | |
6K7F
 
 | |
5BYZ
 
 | | ERK5 in complex with small molecule | | 分子名称: | 4-({5-fluoro-4-[2-methyl-1-(propan-2-yl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)-N-[2-(piperidin-1-yl)ethyl]benzamide, GLYCEROL, Mitogen-activated protein kinase 7 | | 著者 | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | 登録日 | 2015-06-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6GYU
 
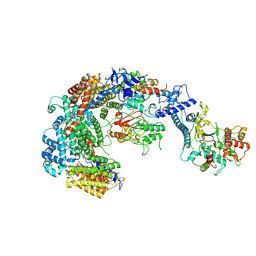 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
