5YHQ
 
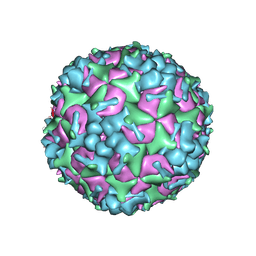 | | Cryo-EM Structure of CVA6 VLP | | 分子名称: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | 著者 | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | 登録日 | 2017-09-29 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | 分子名称: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | 著者 | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | 登録日 | 2017-06-12 | | 公開日 | 2017-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7ZHF
 
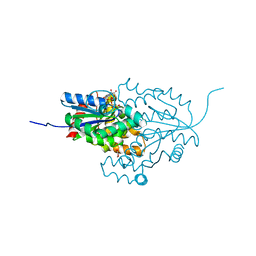 | |
7ZHK
 
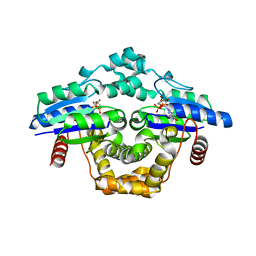 | |
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | 分子名称: | CHLORIDE ION, mBaoJin | | 著者 | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
4O8S
 
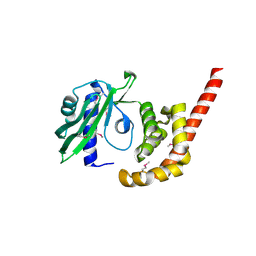 | |
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | 分子名称: | CHLORIDE ION, SULFATE ION, mBaoJin | | 著者 | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | 登録日 | 2023-08-28 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | 分子名称: | CHLORIDE ION, mBaoJin | | 著者 | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | 登録日 | 2023-08-15 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
7KBB
 
 | |
7KBA
 
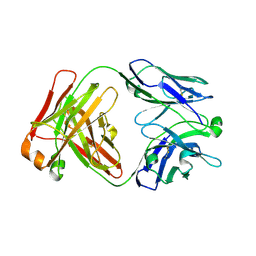 | |
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Li, S, Zheng, Q. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
8UN5
 
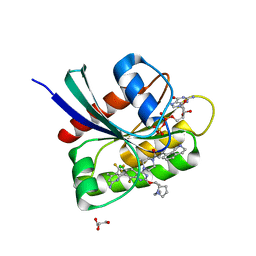 | | KRAS-G13D-GDP in complex with Cpd38 ((E)-1-((3S)-4-(7-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-6-chloro-8-fluoro-2-(((S)-2-methylenetetrahydro-1H-pyrrolizin-7a(5H)-yl)methoxy)quinazolin-4-yl)-3-methylpiperazin-1-yl)-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one) | | 分子名称: | (2E)-1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(4R,7aS)-2-methylidenetetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one, GLYCEROL, GTPase KRas, ... | | 著者 | Ultsch, M.H. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8UN3
 
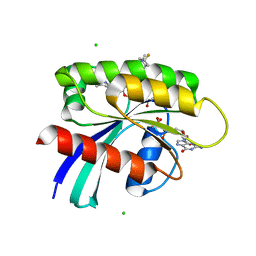 | | KRAS-G13D-GDP in complex with Cpd5 (1-((S)-10-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-11-chloro-7-(((2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl)methoxy)-3,4,13,13a-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-2(1H)-yl)prop-2-en-1-one) | | 分子名称: | 1,2-ETHANEDIOL, 1-[(5M,8aS,13R)-5-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-2-{[(2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl]methoxy}-8a,9,11,12-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-10(8H)-yl]prop-2-en-1-one, CHLORIDE ION, ... | | 著者 | Ultsch, M.H. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8UN4
 
 | |
9B94
 
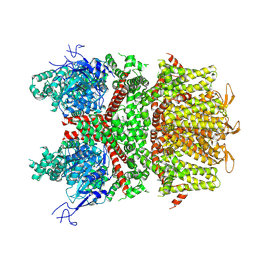 | |
9B91
 
 | |
9B90
 
 | |
9B93
 
 | |
7W6L
 
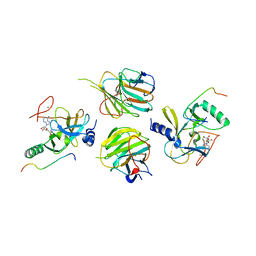 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
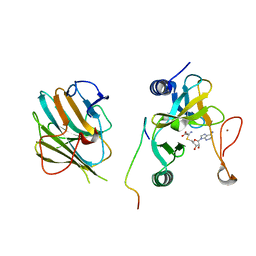 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
9B8Z
 
 | |
9B92
 
 | |
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
