4MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | 分子名称: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | 著者 | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | 登録日 | 1999-03-29 | | 公開日 | 1999-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE AT 1.9 ANGSTROM RESOLUTION | | 分子名称: | COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | 著者 | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | 登録日 | 1999-03-29 | | 公開日 | 1999-06-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
7ROV
 
 | |
6JPV
 
 | |
6K39
 
 | |
8W1L
 
 | |
3KX9
 
 | |
8F6O
 
 | | anti-BTLA monoclonal antibody h22B3 in complex with BTLA | | 分子名称: | B- and T-lymphocyte attenuator, h22B3 Fab heavy chain, h22B3 Fab light chain | | 著者 | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F60
 
 | | anti-BTLA monoclonal antibody r23C8 in complex with BTLA | | 分子名称: | ACETATE ION, B- and T-lymphocyte attenuator, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | 登録日 | 2022-11-15 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6L
 
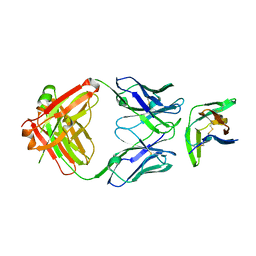 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | 分子名称: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | 著者 | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
3NB9
 
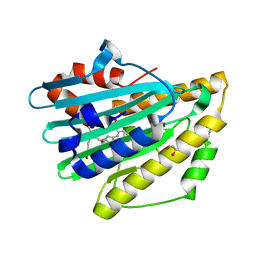 | | Crystal structure of radical H88Q Synechocystis sp. PCYA | | 分子名称: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | 著者 | Gae, D.D. | | 登録日 | 2010-06-03 | | 公開日 | 2010-10-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3NB8
 
 | | Crystal structure of oxidized H88Q Synechocystis sp. PCYA | | 分子名称: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | 著者 | Gae, D.D. | | 登録日 | 2010-06-03 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
6AAW
 
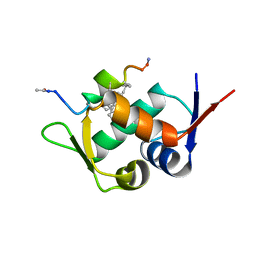 | | Mdm2 in complex with a D amino Acid Containing Stapled Peptide | | 分子名称: | ACE-LEU-THR-PHE-STQ-GLU-TYR-DTR-GLN-LEU-CBA-MK8-SER-ALA-ALA, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Brown, C.J, Partridge, A.W. | | 登録日 | 2018-07-19 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Model System to Explore Macrocyclic Peptide Structural and Chirality Relationships: Tolerance of helix-breaking residues within the context of a stapled peptide.
To Be Published
|
|
3F0M
 
 | |
3F0L
 
 | |
7VWX
 
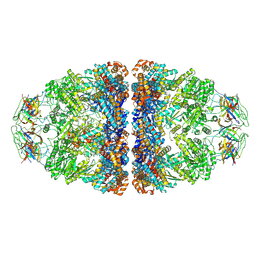 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | 分子名称: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | 著者 | Kim, H, Roh, S.H. | | 登録日 | 2021-11-12 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
7E65
 
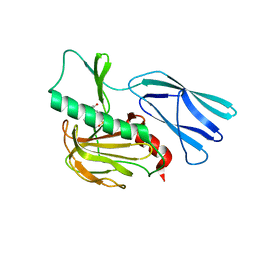 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | 分子名称: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E60
 
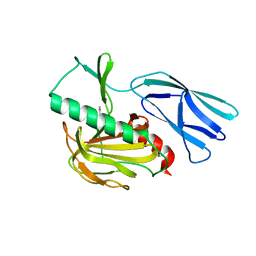 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | 分子名称: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E69
 
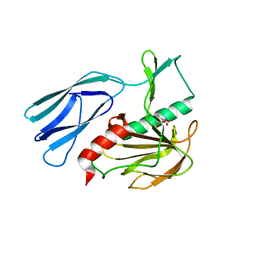 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | 分子名称: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
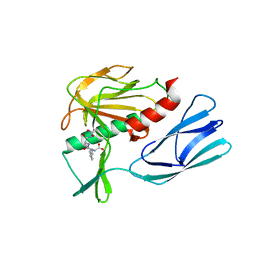 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E64
 
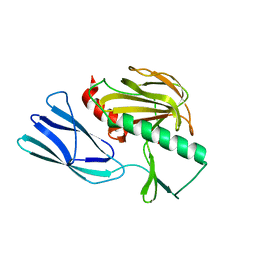 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
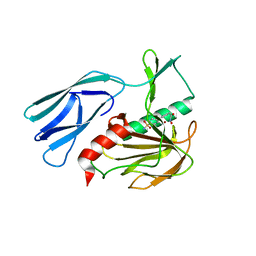 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | 分子名称: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
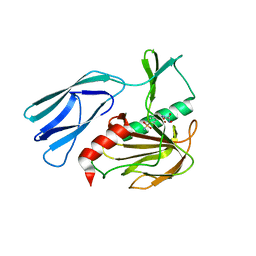 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | 分子名称: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E63
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-1 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(cyclopentylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
8RQU
 
 | | Structure of TEM1 beta-lactamase variant 70.a | | 分子名称: | Beta-lactamase TEM-1, MAGNESIUM ION | | 著者 | Napie, E, Fram, B.F, Gauthier, N.P, Sander, C, Khan, A.R. | | 登録日 | 2024-01-19 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Biorxiv, 2023
|
|
