7XSV
 
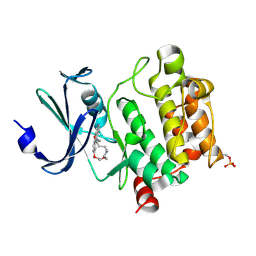 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | 分子名称: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | 著者 | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | 登録日 | 2022-05-15 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
6WO0
 
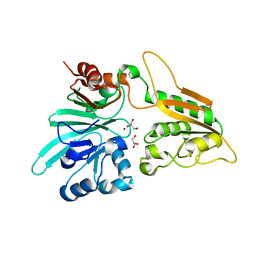 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | 分子名称: | GLYCEROL, Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-23 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WNL
 
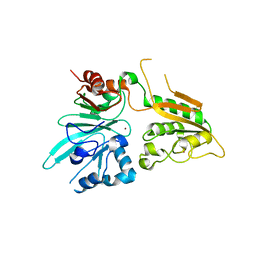 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | 分子名称: | Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8KH5
 
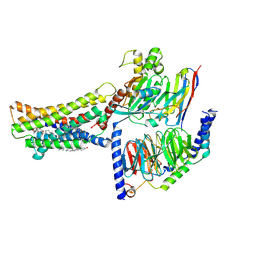 | | Cryo-EM structure of the GPR174-Gs complex bound to endogenous lysoPS | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | 登録日 | 2023-08-21 | | 公開日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
8KGK
 
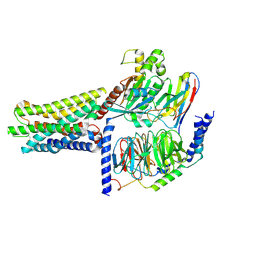 | | Cryo-EM structure of the GPR61-Gs complex | | 分子名称: | G-protein coupled receptor 61, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Nie, Y, Qiu, Z, Zheng, S. | | 登録日 | 2023-08-19 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
8KH4
 
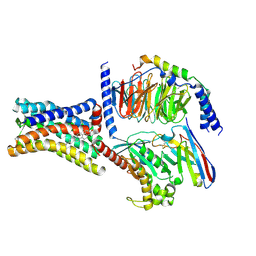 | | Cryo-EM structure of the GPR161-Gs complex | | 分子名称: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | 登録日 | 2023-08-21 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
6MNY
 
 | |
4R29
 
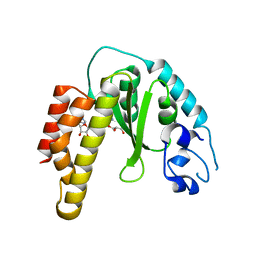 | | Crystal structure of bacterial cysteine methyltransferase effector NleE | | 分子名称: | CITRIC ACID, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | 著者 | Yao, Q, Chen, J, Hu, L, Zhang, L, Shao, F. | | 登録日 | 2014-08-11 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structure and Specificity of the Bacterial Cysteine Methyltransferase Effector NleE Suggests a Novel Substrate in Human DNA Repair Pathway.
Plos Pathog., 10, 2014
|
|
4RAP
 
 | | Crystal structure of bacterial iron-containing dodecameric glycosyltransferase TibC from enterotoxigenic E.coli H10407 | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase TibC | | 著者 | Yao, Q, Lu, Q, Xu, Y, Shao, F. | | 登録日 | 2014-09-10 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.881 Å) | | 主引用文献 | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family.
Elife, 3, 2014
|
|
4RB4
 
 | | Crystal structure of dodecameric iron-containing heptosyltransferase TibC in complex with ADP-D-beta-D-heptose at 3.9 angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase tibC, ... | | 著者 | Yao, Q, Lu, Q, Shao, F. | | 登録日 | 2014-09-12 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.88 Å) | | 主引用文献 | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family
Elife, 3, 2014
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | 分子名称: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | 分子名称: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG1
 
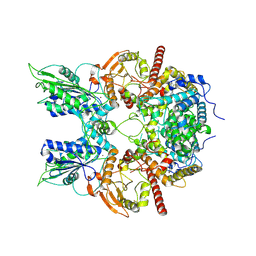 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
6JQH
 
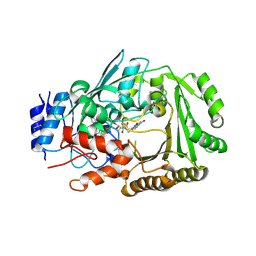 | | Crystal structure of MaDA | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | 著者 | Du, X.X, Lei, X.G. | | 登録日 | 2019-03-31 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
3EIT
 
 | |
3E27
 
 | |
3EIR
 
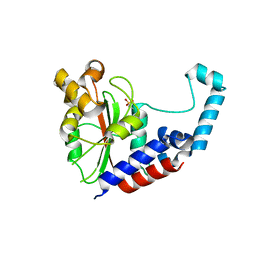 | |
6A73
 
 | | Complex structure of CSN2 with IP6 | | 分子名称: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | 著者 | Liu, L, Li, D, Rao, F, Wang, T. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.447 Å) | | 主引用文献 | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5GZA
 
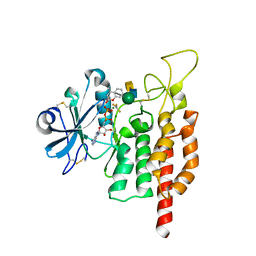 | | protein O-mannose kinase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 4-METHYL-2H-CHROMEN-2-ONE, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Xiao, J. | | 登録日 | 2016-09-27 | | 公開日 | 2016-12-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of protein O-mannose kinase reveals a unique active site architecture
Elife, 5, 2016
|
|
5GUF
 
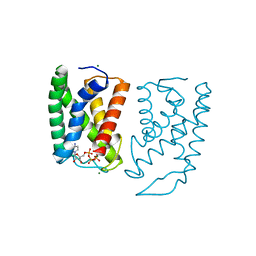 | |
4HCP
 
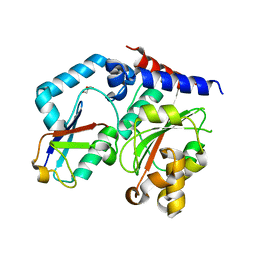 | |
4HCN
 
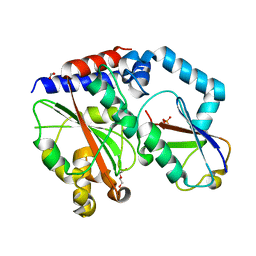 | | Crystal structure of Burkholderia pseudomallei effector protein CHBP in complex with ubiquitin | | 分子名称: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, PHOSPHATE ION, ... | | 著者 | Yao, Q, Cui, J, Zhu, Y, Shao, F. | | 登録日 | 2012-09-30 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7W6L
 
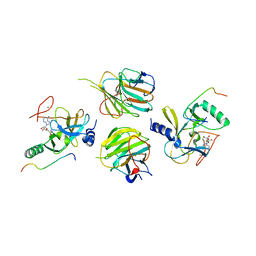 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
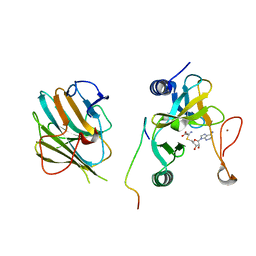 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
