7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-21 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
6LKD
 
 | | in meso full-length rat KMO in complex with a pyrazoyl benzoic acid inhibitor | | 分子名称: | 5-[5-(4-chloranyl-3-fluoranyl-phenyl)-4-methyl-pyrazol-1-yl]-2-phenylmethoxy-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Mimasu, S, Yamagishi, H, Kiyohara, M, Kakefuda, K, Okuda, T. | | 登録日 | 2019-12-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
7DFG
 
 | |
7DFH
 
 | |
8DT8
 
 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2022-07-25 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QPL
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | 登録日 | 2019-02-14 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
8KG5
 
 | | Prefusion RSV F Bound to Lonafarnib and D25 Fab | | 分子名称: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, D25 heavy chain, D25 light chain, ... | | 著者 | Yang, Q, Xue, B, Liu, F, Peng, W, Chen, X. | | 登録日 | 2023-08-17 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.
Signal Transduct Target Ther, 9, 2024
|
|
3K8S
 
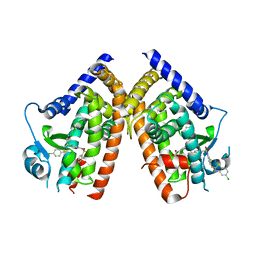 | | Crystal Structure of PPARg in complex with T2384 | | 分子名称: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | 著者 | Wang, Z. | | 登録日 | 2009-10-14 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
8XQR
 
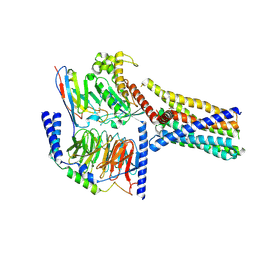 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | 分子名称: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQO
 
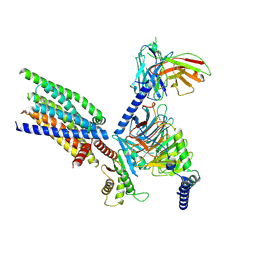 | | Structure of human class T GPCR TAS2R14-Gi complex with Aristolochic acid A. | | 分子名称: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQS
 
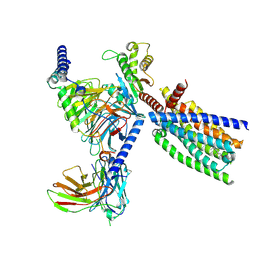 | | Structure of human class T GPCR TAS2R14-DNGi complex with Flufenamic acid. | | 分子名称: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQT
 
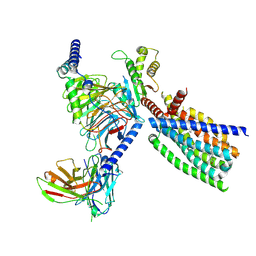 | | Structure of human class T GPCR TAS2R14-Gi complex. | | 分子名称: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQL
 
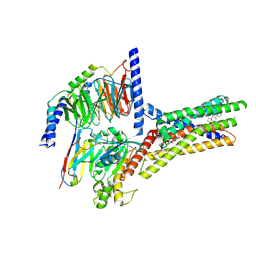 | | Structure of human class T GPCR TAS2R14-miniGs/gust complex with Aristolochic acid A. | | 分子名称: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQP
 
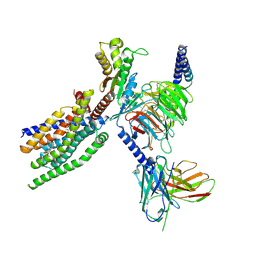 | | Structure of human class T GPCR TAS2R14-Gustducin complex with Aristolochic acid A. | | 分子名称: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8YKY
 
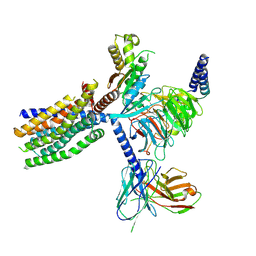 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | 分子名称: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-03-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQN
 
 | | Structure of human class T GPCR TAS2R14-DNGi complex with Aristolochic acid A. | | 分子名称: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | 著者 | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | 登録日 | 2024-01-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
7KXT
 
 | | Crystal structure of human EED | | 分子名称: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | 著者 | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-12-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | 分子名称: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | 著者 | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | 登録日 | 2019-07-22 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (9.83 Å) | | 主引用文献 | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
8XVH
 
 | | Cryo-EM structure of ETBR bound with Endothelin1 | | 分子名称: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
5HU9
 
 | | Crystal structure of ABL1 in complex with CHMFL-074 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]oxy}phenyl)-3-(trifluoromethyl)benzamide, CHLORIDE ION, ... | | 著者 | Kong, L.L, Yun, C.H. | | 登録日 | 2016-01-27 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.529 Å) | | 主引用文献 | Discovery and characterization of a novel potent type II native and mutant BCR-ABL inhibitor (CHMFL-074) for Chronic Myeloid Leukemia (CML)
Oncotarget, 7, 2016
|
|
2HFD
 
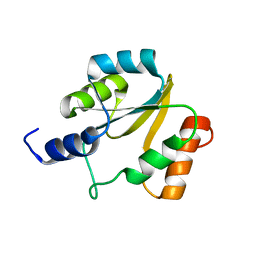 | | NMR structure of protein Hydrogenase-1 operon protein hyaE from Escherichia coli: Northeast Structural Genomics Consortium Target ER415 | | 分子名称: | Hydrogenase-1 operon protein hyaE | | 著者 | Singarapu, K.K, Liu, G, Eletsky, A, Parish, D, Atreya, H.S, Xu, D, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-06-23 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
1D9K
 
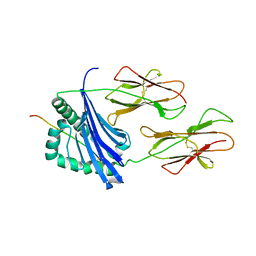 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | 著者 | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | 登録日 | 1999-10-28 | | 公開日 | 1999-12-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
3EVX
 
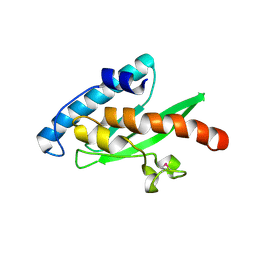 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | 分子名称: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | 著者 | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-10-13 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-01-04 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
8XVJ
 
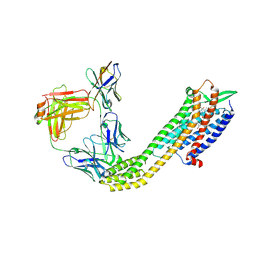 | | Cryo-EM structure of ETAR bound with Macitentan | | 分子名称: | 6-[2-(5-bromanylpyrimidin-2-yl)oxyethoxy]-5-(4-bromophenyl)-~{N}-(propylsulfamoyl)pyrimidin-4-amine, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
 | | Cryo-EM structure of ETAR bound with Ambrisentan | | 分子名称: | (2~{S})-2-(4,6-dimethylpyrimidin-2-yl)oxy-3-methoxy-3,3-diphenyl-propanoic acid, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
