8EHW
 
 | |
8EHX
 
 | |
8P1U
 
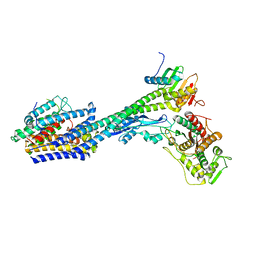 | | Structure of divisome complex FtsWIQLB | | 分子名称: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | 著者 | Yang, L, Chang, S, Tang, D, Dong, H. | | 登録日 | 2023-05-12 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Wang, X, Ma, L, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K8I
 
 | | Crystal structure of Arabidopsis thaliana CRY2 | | 分子名称: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Ma, L, Wang, X, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7U6R
 
 | |
2GM1
 
 | | Crystal structure of the mitotic kinesin eg5 in complex with mg-adp and n-(3-aminopropyl)-n-((3-benzyl-5-chloro-4-oxo-3,4-dihydropyrrolo[2,1-f][1,2,4]triazin-2-yl)(cyclopropyl)methyl)-4-methylbenzamide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-RELATED MOTOR PROTEIN EG5, MAGNESIUM ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2006-04-05 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis and SAR of pyrrolotriazine-4-one based Eg5 inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5CDM
 
 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | 分子名称: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDN
 
 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDQ
 
 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | 分子名称: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDO
 
 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | 分子名称: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5YHQ
 
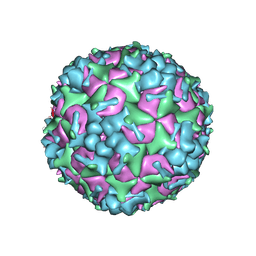 | | Cryo-EM Structure of CVA6 VLP | | 分子名称: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | 著者 | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | 登録日 | 2017-09-29 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
6EF8
 
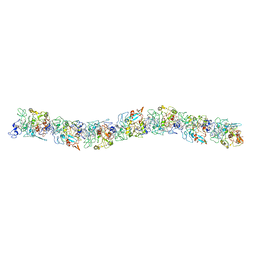 | | Cryo-EM of the OmcS nanowires from Geobacter sulfurreducens | | 分子名称: | C-type cytochrome OmcS, HEME C | | 著者 | Wang, F, Gu, Y, Egelman, E.H, Malvankar, N.S. | | 登録日 | 2018-08-16 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Cell, 177, 2019
|
|
3HPK
 
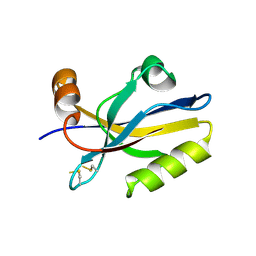 | |
3HPM
 
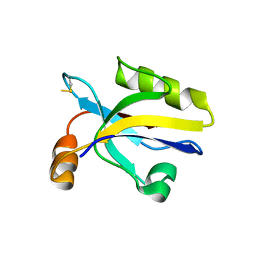 | |
6BFN
 
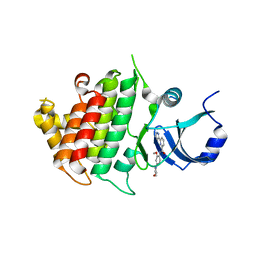 | | Crystal structure of human IRAK1 | | 分子名称: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | 著者 | Wang, L, Qiao, Q, Wu, H. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2ND0
 
 | |
2NCZ
 
 | |
2ND1
 
 | |
9EQG
 
 | |
5XJS
 
 | |
5XJR
 
 | |
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | 著者 | Ting, Y.T, Broury, A, Ooi, J. | | 登録日 | 2023-06-29 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.12621117 Å) | | 主引用文献 | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
