6DJP
 
 | | Integrin alpha-v beta-8 in complex with the Fabs 8B8 and 68 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 68 heavy chain Fab, ... | | 著者 | Cormier, A, Campbell, M.G, Nishimura, S.L, Cheng, Y. | | 登録日 | 2018-05-25 | | 公開日 | 2018-07-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Cryo-EM structure of the alpha v beta 8 integrin reveals a mechanism for stabilizing integrin extension.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6XHN
 
 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | 分子名称: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.377 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHM
 
 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.406 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
4GA7
 
 | | Crystal structure of human serpinB1 mutant | | 分子名称: | Leukocyte elastase inhibitor | | 著者 | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | 登録日 | 2012-07-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
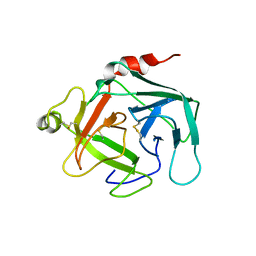 | | Crystal structure of active human granzyme H | | 分子名称: | CHLORIDE ION, Granzyme H, SULFATE ION | | 著者 | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | 登録日 | 2012-07-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
5GNU
 
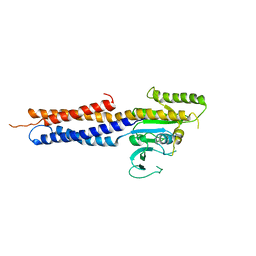 | | the structure of mini-MFN1 apo | | 分子名称: | Mitofusin-1 | | 著者 | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | 登録日 | 2016-07-25 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.113 Å) | | 主引用文献 | BDLP-like folding of Mitofusin 1
To Be Published
|
|
8HNW
 
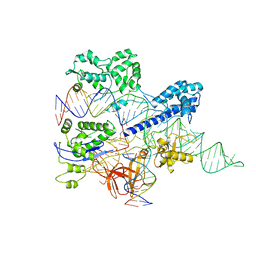 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | 著者 | Sun, W, Cheng, Z, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | 分子名称: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | 著者 | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
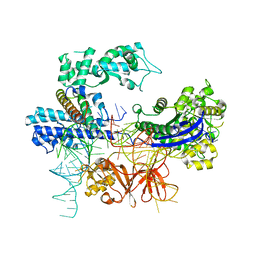 | |
8HNV
 
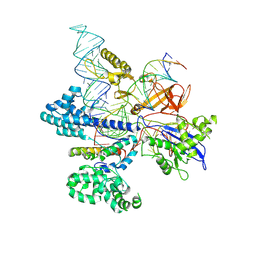 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | 分子名称: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | 著者 | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6AGG
 
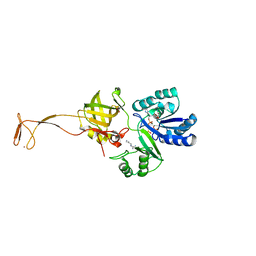 | |
7ROJ
 
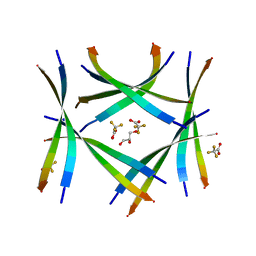 | | Amyloid-related segment of alphaB-crystallin residues 90-100 with G95W mutation | | 分子名称: | Alpha-crystallin B chain peptide, GLYCEROL, SODIUM ION, ... | | 著者 | Sawaya, M.R, Do, T.D, Eisenberg, D.S. | | 登録日 | 2021-07-30 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Atomic view of an amyloid dodecamer exhibiting selective cellular toxic vulnerability in acute brain slices.
Protein Sci., 31, 2022
|
|
7ROL
 
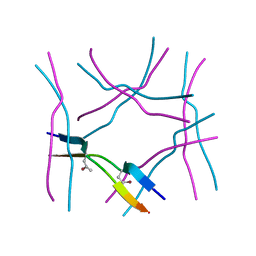 | |
8AYS
 
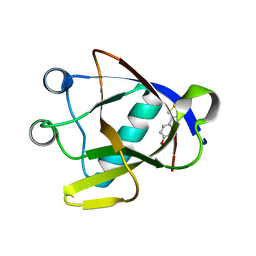 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | 分子名称: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | 登録日 | 2022-09-03 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8AZ8
 
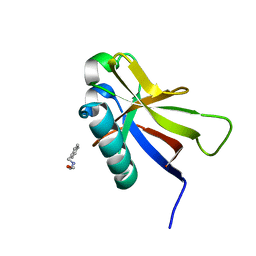 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | 分子名称: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | 登録日 | 2022-09-05 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8A55
 
 | |
3JUE
 
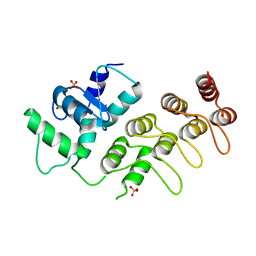 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | 分子名称: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | 著者 | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | 登録日 | 2009-09-15 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
4KK1
 
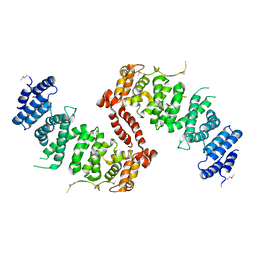 | | Crystal Structure of TSC1 core domain from S. pombe | | 分子名称: | Tuberous sclerosis 1 protein homolog | | 著者 | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | 登録日 | 2013-05-05 | | 公開日 | 2013-07-17 | | 最終更新日 | 2013-07-31 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
4KK0
 
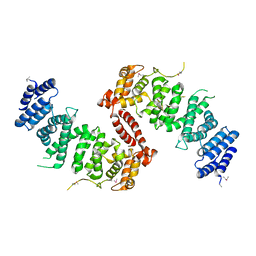 | | Crystal Structure of TSC1 core domain from S. pombe | | 分子名称: | Tuberous sclerosis 1 protein homolog | | 著者 | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | 登録日 | 2013-05-05 | | 公開日 | 2013-07-17 | | 最終更新日 | 2013-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | 分子名称: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
6XHL
 
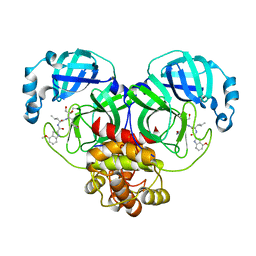 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 分子名称: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.471 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHO
 
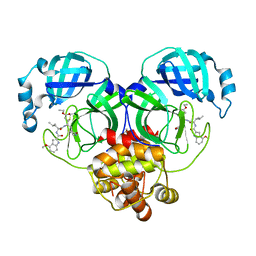 | | Covalent complex of SARS-CoV main protease with ethyl (4R)-4-({N-[(4-methoxy-1H-indol-2-yl)carbonyl]-L-leucyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.446 Å) | | 主引用文献 | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
8CRF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | 分子名称: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | 著者 | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | 登録日 | 2023-03-08 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRK
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | 分子名称: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | 登録日 | 2023-03-08 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
