7T50
 
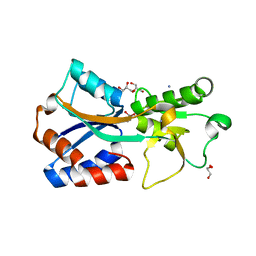 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in chromate-bound form | | 分子名称: | AMMONIUM ION, Chromate, GLYCEROL, ... | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T51
 
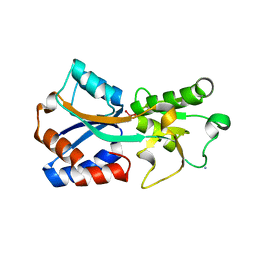 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in molybdate-bound form | | 分子名称: | AMMONIUM ION, GLYCEROL, MOLYBDATE ION, ... | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
8HTW
 
 | |
5F8T
 
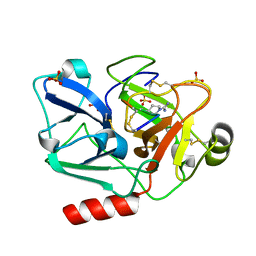 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-2 | | 分子名称: | CYS-PRO-LYS-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein light chain, SULFATE ION, ... | | 著者 | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-2
To Be Published
|
|
5F8Z
 
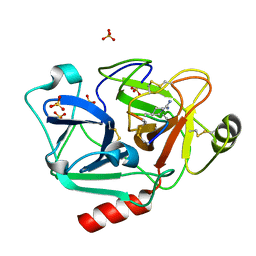 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | 分子名称: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | 著者 | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
5F8X
 
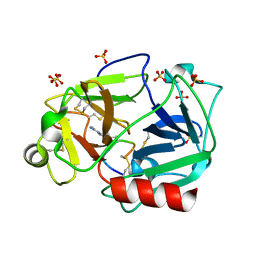 | | The crystal structure of human plasma kallikrein in complex with its peptide inhibitor pkalin-3 | | 分子名称: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-TRP-CYS, Plasma kallikrein, SULFATE ION, ... | | 著者 | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Crystal Structure Of Human Plasma Kallikrein In Complex With Its Peptide Inhibitor Pkalin-3
To Be Published
|
|
6A6B
 
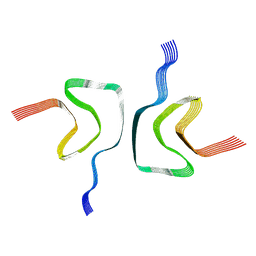 | | cryo-em structure of alpha-synuclein fiber | | 分子名称: | Alpha-synuclein | | 著者 | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | 登録日 | 2018-06-27 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
4IW2
 
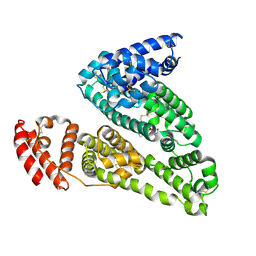 | | HSA-glucose complex | | 分子名称: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | 著者 | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | 登録日 | 2013-01-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4K2C
 
 | | HSA Ligand Free | | 分子名称: | Serum albumin | | 著者 | Wang, Y, Luo, Z, Shi, X, Huang, M. | | 登録日 | 2013-04-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2018-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
7LVP
 
 | | Cryptococcus neoformans AIR synthetase | | 分子名称: | 1,2-ETHANEDIOL, AIR synthase, GLYCINE, ... | | 著者 | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
7LVO
 
 | | Cryptococcus neoformans GAR synthetase | | 分子名称: | 1,2-ETHANEDIOL, phosphoribosyl-glycinamide (GAR) synthetase | | 著者 | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
7L5S
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | 著者 | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | 登録日 | 2020-12-22 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | 登録日 | 2020-12-22 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.733 Å) | | 主引用文献 | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | 分子名称: | NAD(+) hydrolase SARM1 | | 著者 | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | 分子名称: | Isoform B of NAD(+) hydrolase sarm1 | | 著者 | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-04-21 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCZ
 
 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | 分子名称: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | 著者 | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7M6K
 
 | |
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | 分子名称: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | 著者 | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | 登録日 | 2021-04-13 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
5ZM6
 
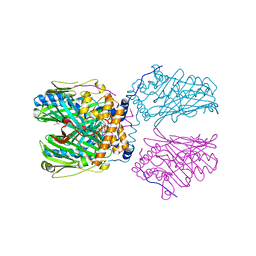 | | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 | | 分子名称: | ACETATE ION, Oxysterol-binding protein-related protein 1, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | 著者 | Dong, J, Wang, J, Luo, Z, Wu, J.W. | | 登録日 | 2018-04-01 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
7ECH
 
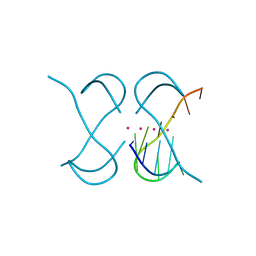 | | Crystal Structure of d(G4C2)2-K in F222 space group | | 分子名称: | DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3'), POTASSIUM ION | | 著者 | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | 登録日 | 2021-03-12 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
7ECG
 
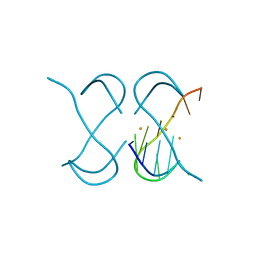 | | Crystal Structure of d(G4C2)2-Ba in F222 space group | | 分子名称: | BARIUM ION, DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3') | | 著者 | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | 登録日 | 2021-03-12 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
7ECF
 
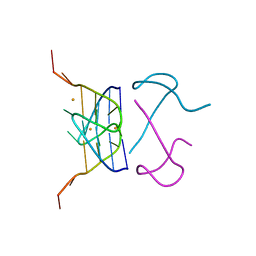 | | Crystal Structure of d(G4C2)2-Ba in C2221 space group | | 分子名称: | BARIUM ION, DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3') | | 著者 | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | 登録日 | 2021-03-12 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFT
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | 分子名称: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFW
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
