3WRO
 
 | |
3WRR
 
 | |
6ISU
 
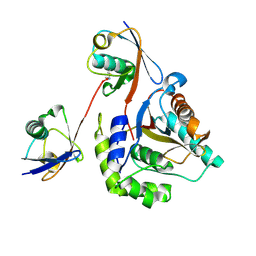 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | 分子名称: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | 著者 | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | 登録日 | 2018-11-19 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3WRQ
 
 | |
3WRN
 
 | |
3WRS
 
 | |
4PP4
 
 | |
8GMU
 
 | |
8GMT
 
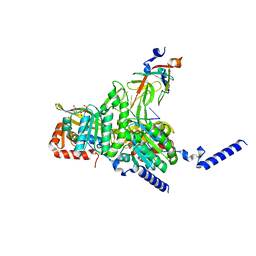 | | Structure of UmuD in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA polymerase V subunit UmuD, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMS
 
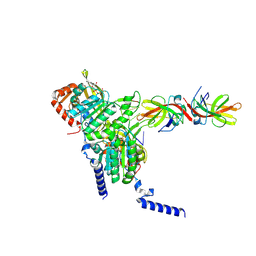 | | Structure of LexA in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UP2
 
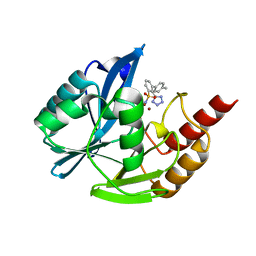 | | NDM1-inhibitor co-structure | | 分子名称: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | 著者 | Scapin, G, Fischmann, T.O. | | 登録日 | 2022-04-14 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
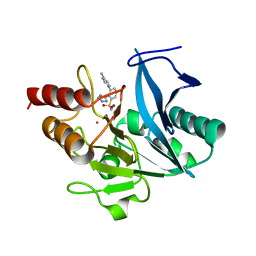 | | NDM1-inhibitor co-structure | | 分子名称: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | 著者 | Scapin, G, Fischmann, T.O. | | 登録日 | 2022-04-14 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
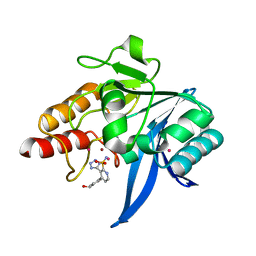 | | NDM1-inhibitor co-structure | | 分子名称: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | 著者 | Scapin, G, Fischmann, T.O. | | 登録日 | 2022-04-14 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
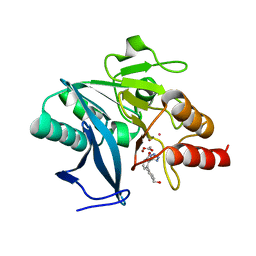 | | NDM1-inhibitor co-structure | | 分子名称: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | 著者 | Scapin, G, Fischmann, T.O. | | 登録日 | 2022-04-14 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
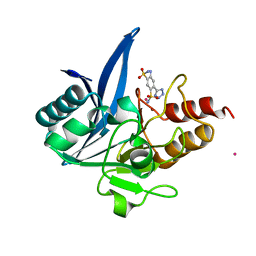 | | NDM1-inhibitor co-structure | | 分子名称: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | 著者 | Scapin, G, Fischmann, T.O. | | 登録日 | 2022-04-14 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
5C5Q
 
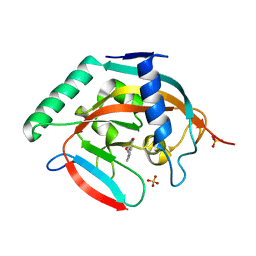 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5R
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (7R)-2-hydroxy-7-(propan-2-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine-3-carbonitrile, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2RHU
 
 | |
2RI7
 
 | |
2RI5
 
 | |
2RI2
 
 | |
3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | 分子名称: | CAFFEINE, Chitinase | | 著者 | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | 登録日 | 2009-02-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
6JX4
 
 | |
