5CEV
 
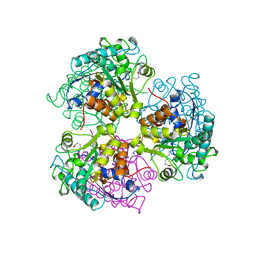 | | ARGINASE FROM BACILLUS CALDEVELOX, L-LYSINE COMPLEX | | 分子名称: | GUANIDINE, LYSINE, MANGANESE (II) ION, ... | | 著者 | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | 登録日 | 1999-03-16 | | 公開日 | 1999-04-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
8DIT
 
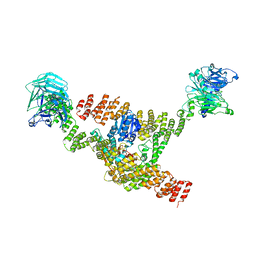 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | 分子名称: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | 著者 | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | 登録日 | 2022-06-29 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
2IE3
 
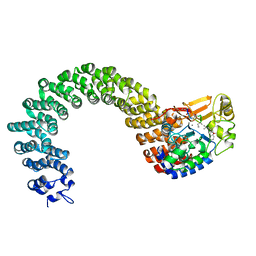 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to Tumor-inducing Toxins | | 分子名称: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | 著者 | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | 登録日 | 2006-09-17 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
2IE4
 
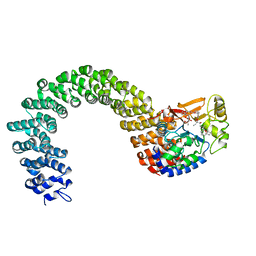 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | 分子名称: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | 著者 | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | 登録日 | 2006-09-17 | | 公開日 | 2006-11-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
2HV6
 
 | |
5EP2
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain in Complex with AzaU Inhibitor | | 分子名称: | 2,2-dimethylpropyl 2-[[3,5-bis(oxidanylidene)-2~{H}-1,2,4-triazin-6-yl]sulfanyl]ethanoate, ACETATE ION, Putative repressor protein luxO | | 著者 | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2015-11-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.421 Å) | | 主引用文献 | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP1
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | 分子名称: | ACETATE ION, Putative repressor protein luxO | | 著者 | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2015-11-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP4
 
 | | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2015-11-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP3
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain Bound to CV-133 Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2,2-dimethylpropyl 2-[(3-oxidanylidene-5-sulfanylidene-2~{H}-1,2,4-triazin-6-yl)amino]ethanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2015-11-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP0
 
 | | Quorum-Sensing Signal Integrator LuxO - Receiver+Catalytic Domains | | 分子名称: | 1,2-ETHANEDIOL, Putative repressor protein luxO, SULFATE ION | | 著者 | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2015-11-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
2CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, NATIVE STRUCTURE AT PH 8.5 | | 分子名称: | GUANIDINE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | 著者 | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | 登録日 | 1999-03-10 | | 公開日 | 1999-04-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
2F1W
 
 | | Crystal structure of the TRAF-like domain of HAUSP/USP7 | | 分子名称: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | 登録日 | 2005-11-15 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
2F1S
 
 | |
2F1Z
 
 | | Crystal structure of HAUSP | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | 登録日 | 2005-11-15 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
2F1Y
 
 | |
2F1X
 
 | |
1FBV
 
 | | STRUCTURE OF A CBL-UBCH7 COMPLEX: RING DOMAIN FUNCTION IN UBIQUITIN-PROTEIN LIGASES | | 分子名称: | SIGNAL TRANSDUCTION PROTEIN CBL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E12-18 KDA UBCH7, ... | | 著者 | Zheng, N, Wang, P, Jeffrey, P.D, Pavletich, N.P. | | 登録日 | 2000-07-17 | | 公開日 | 2000-08-30 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases.
Cell(Cambridge,Mass.), 102, 2000
|
|
1FQV
 
 | | Insights into scf ubiquitin ligases from the structure of the skp1-skp2 complex | | 分子名称: | SKP1, SKP2 | | 著者 | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | 登録日 | 2000-09-06 | | 公開日 | 2000-11-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
1FS2
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | 分子名称: | SKP1, SKP2 | | 著者 | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | 登録日 | 2000-09-08 | | 公開日 | 2000-11-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
1FS1
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | 分子名称: | CYCLIN A/CDK2-ASSOCIATED P19, CYCLIN A/CDK2-ASSOCIATED P45 | | 著者 | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | 登録日 | 2000-09-08 | | 公開日 | 2000-11-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
4M9R
 
 | |
4L9O
 
 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2013-06-18 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
4KMO
 
 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | 分子名称: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | 著者 | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | 登録日 | 2013-05-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
4H5J
 
 | |
4H5I
 
 | |
