6FOY
 
 | |
6FP0
 
 | |
6FOX
 
 | |
6RPP
 
 | | Crystal structure of PabCDC21-1 intein | | 分子名称: | ACETATE ION, Cell division control protein, DI(HYDROXYETHYL)ETHER | | 著者 | Mikula, K.M, Beyer, H.M, Iwai, H. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6RPQ
 
 | | Crystal structure of PhoCDC21-1 intein | | 分子名称: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | 著者 | Beyer, H.M, Mikula, K.M, Iwai, H. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.654 Å) | | 主引用文献 | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | 分子名称: | Redirecting phage packaging protein C (RppC) | | 著者 | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | 登録日 | 2018-09-11 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
6PW8
 
 | |
6XR3
 
 | |
6FP1
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1 | | 分子名称: | 2-(6-chloranyl-5,7-dimethyl-3-oxidanylidene-1,4-benzoxazin-4-yl)ethanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Levy, C.W, Leys, D. | | 登録日 | 2018-02-08 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
6MRX
 
 | | Sialidase26 apo | | 分子名称: | Sialidase26 | | 著者 | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MYV
 
 | | Sialidase26 co-crystallized with DANA-Gc | | 分子名称: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | 著者 | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | 登録日 | 2018-11-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | 分子名称: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | 著者 | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | 登録日 | 2017-12-18 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
7ZJU
 
 | |
6ZNW
 
 | |
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | 分子名称: | Transient receptor potential cation channel subfamily V member 1 | | 著者 | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | 登録日 | 2015-02-02 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.275 Å) | | 主引用文献 | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
3TGP
 
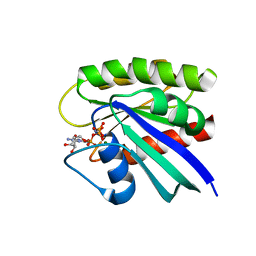 | | Room temperature H-ras | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Fraser, J.S, Alber, T. | | 登録日 | 2011-08-17 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3075 Å) | | 主引用文献 | Accessing protein conformational ensembles using room-temperature X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | 登録日 | 2012-05-29 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.9228 Å) | | 主引用文献 | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
7SOZ
 
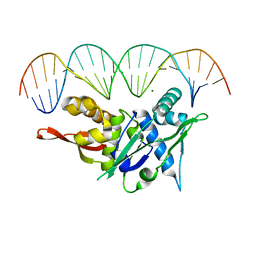 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (5 mg/mL EDC, 12 hours). | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | 著者 | Ward, A.R, Snow, C.D. | | 登録日 | 2021-11-01 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7SPM
 
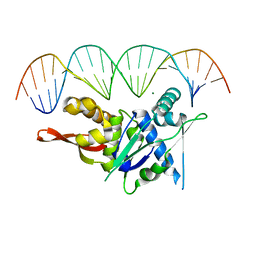 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (30 mg/mL EDC, 12 hours, 2 doses). | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | 著者 | Ward, A.R, Snow, C.D. | | 登録日 | 2021-11-02 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7SQM
 
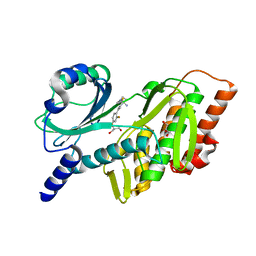 | | Discovery and Preclinical Pharmacology of INE963, A Potent and Fast-Acting Blood-Stage Antimalarial with a High Barrier to Resistance and Potential for Single-Dose Cure in Uncomplicated Malaria | | 分子名称: | 1-[(4S)-5-(2,4-difluorophenyl)imidazo[2,1-b][1,3,4]thiadiazol-2-yl]-4-methylpiperidin-4-amine, GLYCEROL, Serine/threonine-protein kinase haspin | | 著者 | Shu, W, Yokokawa, F. | | 登録日 | 2021-11-05 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Failure of artesunate-mefloquine combination therapy for uncompli-cated Plasmodium falciparum malaria in southern Cambodia.
Malar. J., 2009, 8, 10, 2009
|
|
7LM4
 
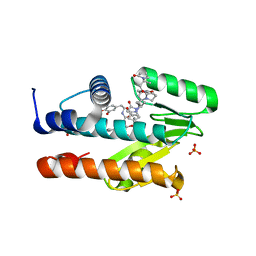 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | 分子名称: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | 著者 | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | 登録日 | 2021-02-05 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8V1J
 
 | |
8GJR
 
 | |
6TYF
 
 | |
6TYG
 
 | |
