8WYG
 
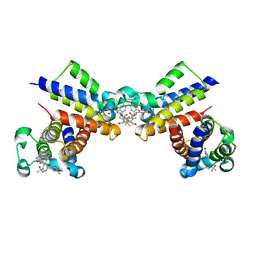 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | 分子名称: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WXY
 
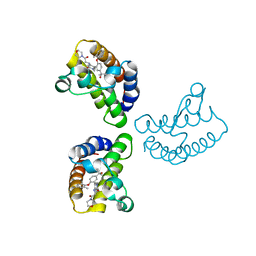 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 23 | | 分子名称: | 5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-4-[(2-morpholin-4-yl-2-oxidanylidene-ethyl)amino]pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
5ZCK
 
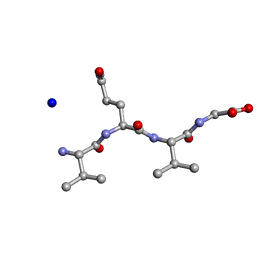 | | Structure of the RIP3 core region | | 分子名称: | SODIUM ION, peptide from Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Li, J, Wu, H. | | 登録日 | 2018-02-18 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.271 Å) | | 主引用文献 | The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex.
Cell, 173, 2018
|
|
4QQ6
 
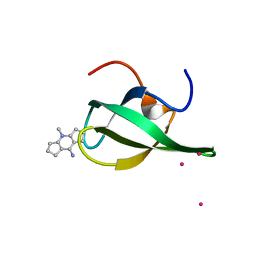 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | 分子名称: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-26 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4QQD
 
 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | 分子名称: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-27 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
3CLY
 
 | |
4HK7
 
 | |
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | 分子名称: | Uracil-5-carboxylate decarboxylase, ZINC ION | | 著者 | Xu, S, Zhu, J, Ding, J. | | 登録日 | 2012-10-15 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
9ERC
 
 | | Hydrogenase-2 Ni-Li state | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | 著者 | Wong, K.L, Carr, S.B, Ash, P.A, Vincent, K.A. | | 登録日 | 2024-03-22 | | 公開日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.308 Å) | | 主引用文献 | Glutamate flick enables proton tunnelling during fast redox biocatalysis
To Be Published
|
|
7CLD
 
 | | Crystal structure of T2R-TTL-Cevipabulin complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, CALCIUM ION, ... | | 著者 | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.611 Å) | | 主引用文献 | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
7DP8
 
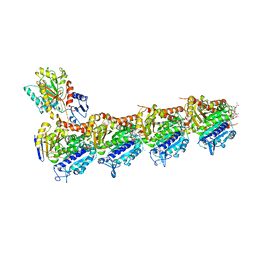 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | 分子名称: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | 著者 | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | 登録日 | 2020-12-18 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.446 Å) | | 主引用文献 | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
8H01
 
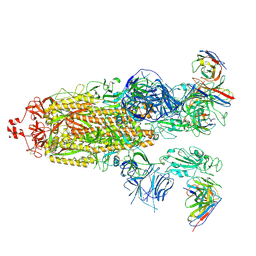 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-09-27 | | 公開日 | 2023-04-12 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
9GCX
 
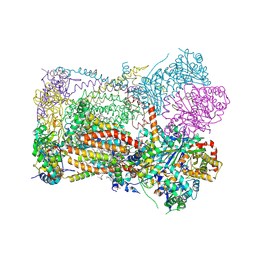 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor F8 | | 分子名称: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, 2-methyl-3-(4-(4-(trifluoromethoxy)phenoxy)phenyl)-1,5,7,8-tetrahydro-4H-pyrano[4,3-b]pyridin-4-one, ... | | 著者 | Pinthong, N, Hong, W, ONeill, P.W, Antonyuk, S.V. | | 登録日 | 2024-08-03 | | 公開日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (3.522 Å) | | 主引用文献 | Novel antimalarial 3-substituted quinolones isosteres with improved pharmacokinetic properties.
Eur.J.Med.Chem., 284, 2024
|
|
4HK6
 
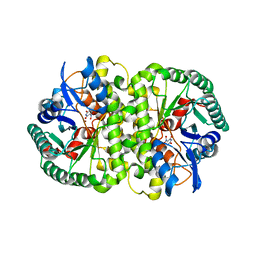 | |
8H00
 
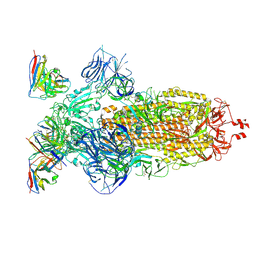 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-09-27 | | 公開日 | 2023-04-12 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8GZZ
 
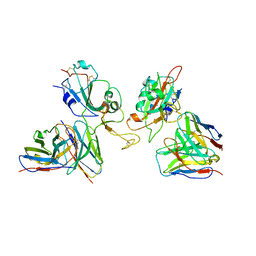 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | 分子名称: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-09-27 | | 公開日 | 2023-04-12 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8J6T
 
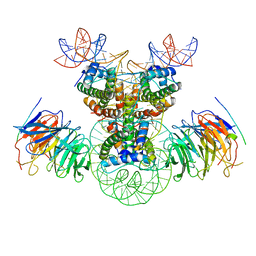 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6S
 
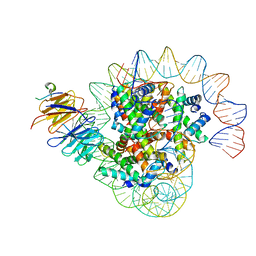 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | 分子名称: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | 著者 | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | 登録日 | 2007-09-24 | | 公開日 | 2008-03-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.1 Å) | | 主引用文献 | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
1P6C
 
 | | crystal structure of phosphotriesterase triple mutant H254G/H257W/L303T complexed with diisopropylmethylphosphonate | | 分子名称: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, METHYLPHOSPHONIC ACID DIISOPROPYL ESTER, Parathion hydrolase, ... | | 著者 | Hill, C.M, Li, W, Thoden, J.B, Holden, H.M, Raushel, F.M. | | 登録日 | 2003-04-29 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site.
J.Am.Chem.Soc., 125, 2003
|
|
1P6B
 
 | | X-ray structure of phosphotriesterase, triple mutant H254G/H257W/L303T | | 分子名称: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, ETHYL DIHYDROGEN PHOSPHATE, Parathion hydrolase, ... | | 著者 | Hill, C.M, Li, W, Thoden, J.B, Holden, H.M, Raushel, F.M. | | 登録日 | 2003-04-29 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site.
J.Am.Chem.Soc., 125, 2003
|
|
4HJW
 
 | |
8IG4
 
 | |
