2E6W
 
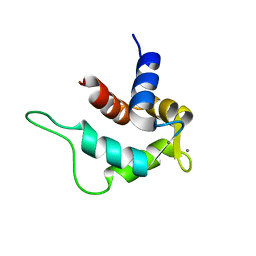 | | Solution structure and calcium binding properties of EF-hands 3 and 4 of calsenilin | | 分子名称: | CALCIUM ION, Calsenilin | | 著者 | Yu, L, Sun, C, Mendoza, R, Hebert, E, Pereda-Lopez, A, Hajduk, P.J, Olejniczak, E.T. | | 登録日 | 2007-01-05 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and calcium-binding properties of EF-hands 3 and 4 of calsenilin.
Protein Sci., 16, 2007
|
|
2E0T
 
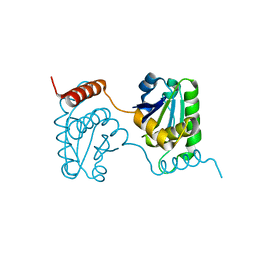 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | 分子名称: | Dual specificity phosphatase 26 | | 著者 | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-10-13 | | 公開日 | 2007-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5JEN
 
 | |
4YFZ
 
 | |
5JFB
 
 | | Crystal structure of the scavenger receptor cysteine-rich domain 5 (SRCR5) from porcine CD163 | | 分子名称: | Scavenger receptor cysteine-rich type 1 protein M130 | | 著者 | Ma, H, Jiang, L, Qiao, S, Zhang, G, Li, R. | | 登録日 | 2016-04-19 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection
J. Virol., 91, 2017
|
|
7T2Y
 
 | | X-ray structure of a designed cold unfolding four helix bundle | | 分子名称: | Designed cold unfolding four helix bundle | | 著者 | Harrison, J.S, Kuhlman, B, Szyperski, T, Premkumar, L, Maguire, J, Pulavarti, S, Yuen, S. | | 登録日 | 2021-12-06 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
1A2A
 
 | | AGKISTROTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | 分子名称: | CHLORIDE ION, PHOSPHOLIPASE A2 | | 著者 | Tang, L, Zhou, Y, Lin, Z. | | 登録日 | 1997-12-25 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of agkistrodotoxin, a phospholipase A2-type presynaptic neurotoxin from agkistrodon halys pallas.
J.Mol.Biol., 282, 1998
|
|
4RLH
 
 | | Crystal structure of enoyl ACP reductase from Burkholderia pseudomallei in complex with AFN-1252 | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | 著者 | Rao, K.N, Sarah, J, Anirudha, L, Subramanya, H.S. | | 登録日 | 2014-10-17 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal structure of enoyl ACP reductase from
Burkholderia pseudomallei in complex with AFN-1252
To be Published
|
|
5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-03-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5J07
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P1/2 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-27 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
4YBH
 
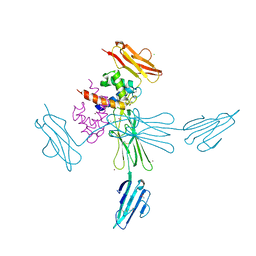 | |
5J0C
 
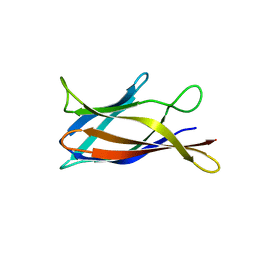 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P2/3 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5JNP
 
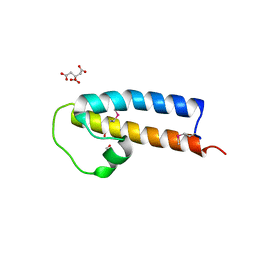 | | Crystal structure of a rice (Oryza Sativa) cellulose synthase plant conserved region (P-CR) | | 分子名称: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Rushton, P.S, Olek, A.T, Makowski, L, Badger, J, Steussy, C.N, Carpita, N.C, Stauffacher, C.V. | | 登録日 | 2016-04-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | Rice Cellulose SynthaseA8 Plant-Conserved Region Is a Coiled-Coil at the Catalytic Core Entrance.
Plant Physiol., 173, 2017
|
|
4YOQ
 
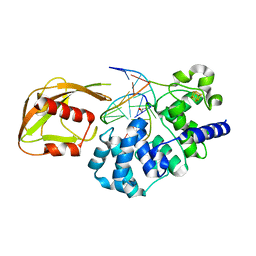 | | Crystal Structure of MutY bound to its anti-substrate | | 分子名称: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | 著者 | Wang, L, Lee, S, Verdine, G.L. | | 登録日 | 2015-03-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
4RKA
 
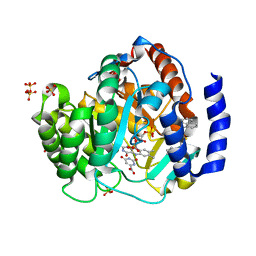 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347 | | 分子名称: | 2-{[5-(naphthalen-1-ylmethyl)-4-oxo-4H-1lambda~4~,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | 著者 | Zhu, L, Ren, X, Zhu, J, Li, H. | | 登録日 | 2014-10-12 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347
TO BE PUBLISHED
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
4RPN
 
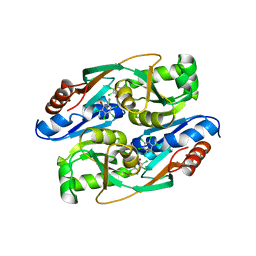 | | PcpR inducer binding domain complex with pentachlorophenol | | 分子名称: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | 著者 | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | 登録日 | 2014-10-30 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4YPQ
 
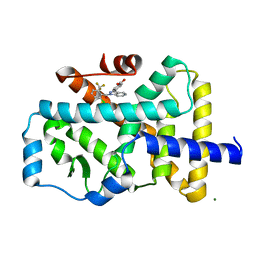 | | Crystal structure of the ROR(gamma)t ligand binding domain in complex with 4-(1-(2-chloro-6-(trifluoromethyl)benzoyl)-1H-indazol-3-yl)benzoic acid | | 分子名称: | 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, MAGNESIUM ION, Nuclear receptor ROR-gamma | | 著者 | Leysen, S, Scheepstra, M, van Almen, G.C, Ottmann, C, Brunsveld, L. | | 登録日 | 2015-03-13 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Identification of an allosteric binding site for ROR gamma t inhibition.
Nat Commun, 6, 2015
|
|
5L42
 
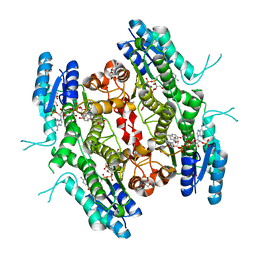 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 3 | | 分子名称: | (2~{R})-2-[3,4-bis(oxidanyl)phenyl]-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | 登録日 | 2016-05-24 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | 分子名称: | Cdc42-interacting protein 4 | | 著者 | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-02-23 | | 公開日 | 2007-05-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
7UIN
 
 | |
4RUS
 
 | | Carp Fishelectin, holo form | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Capaldi, S, Faggion, B, Carrizo, M.E, Destefanis, L, Gonzalez, M.C, Perduca, M, Bovi, M, Galliano, M, Monaco, H.L. | | 登録日 | 2014-11-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Three-dimensional structure and ligand-binding site of carp fishelectin (FEL).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7UIM
 
 | |
4I87
 
 | | Crystal structure of TTR variant I84S in complex with CHF5074 at acidic pH | | 分子名称: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, Transthyretin | | 著者 | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | 登録日 | 2012-12-03 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
4RX2
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | 分子名称: | Beta-lactamase TEM, SULFATE ION | | 著者 | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-12-08 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.315 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
