8DAH
 
 | | [20 bp edge] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | 分子名称: | DNA (5'-D(*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-06-13 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (5.47 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8IW9
 
 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW4
 
 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8WCM
 
 | |
8IW7
 
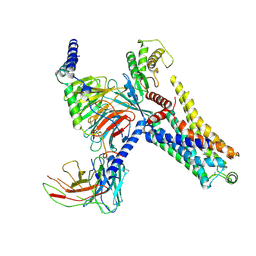 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | 分子名称: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ITF
 
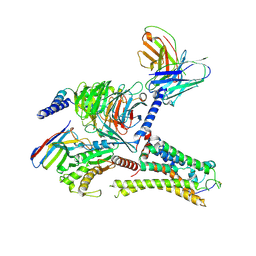 | | Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-22 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
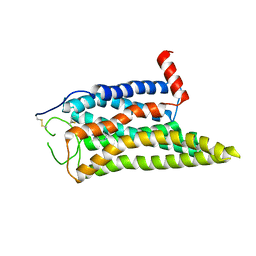 | | Cryo-EM structure of the SPE-mTAAR9 complex | | 分子名称: | SPERMIDINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
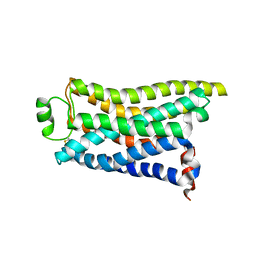 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | 分子名称: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW1
 
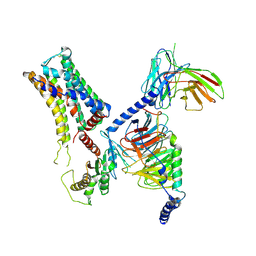 | | Cryo-EM structure of the PEA-bound mTAAR9-Golf complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8CS8
 
 | | [High G:C, High Vapor Diffusion] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*GP*CP*CP*TP*GP*CP*GP*CP*GP*GP*AP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*CP*GP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-05-12 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS4
 
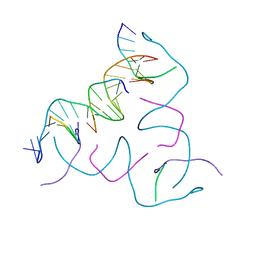 | | [AGC/GTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | 分子名称: | DNA (5'-D(*AP*GP*CP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-05-12 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6.03 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CYN
 
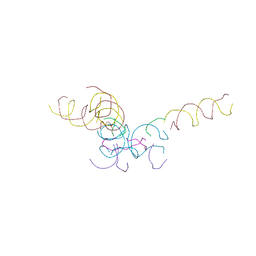 | | [2T7+20bp Linker] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle with 20 bp Sticky-End Linker | | 分子名称: | DNA (5'-D(*GP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-05-24 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (9.45 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS3
 
 | | [CAG/TCG] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-05-12 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (5.14 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS5
 
 | | [GGA/CTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | 分子名称: | DNA (5'-D(*CP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | 登録日 | 2022-05-12 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6.52 Å) | | 主引用文献 | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7JTN
 
 | |
7CRH
 
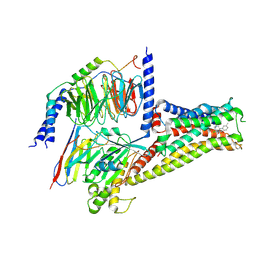 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Yan, W, Shao, Z.H. | | 登録日 | 2020-08-13 | | 公開日 | 2021-03-03 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
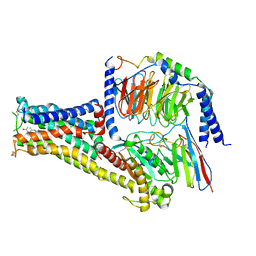 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, W. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
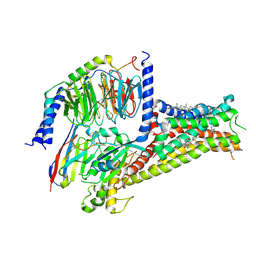 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
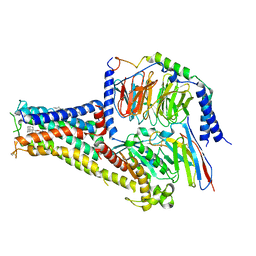 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7KJS
 
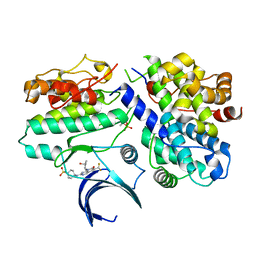 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | 分子名称: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | 著者 | McTigue, M.A, He, Y, Ferre, R.A. | | 登録日 | 2020-10-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7JTQ
 
 | |
7P1K
 
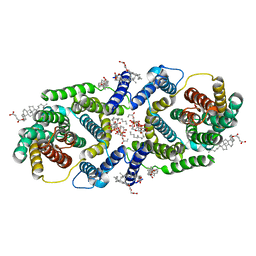 | | Cryo EM structure of bison NHA2 in nano disc structure | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | 著者 | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | 登録日 | 2021-07-01 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1J
 
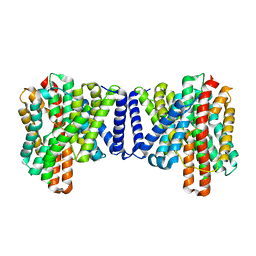 | | Cryo EM structure of bison NHA2 in detergent structure | | 分子名称: | mitochondrial sodium/hydrogen exchanger 9B2 | | 著者 | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | 登録日 | 2021-07-01 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
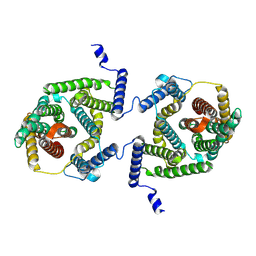 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | 分子名称: | mitochondrial sodium/hydrogen exchanger 9B2 | | 著者 | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | 登録日 | 2021-07-01 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
