2HE2
 
 | | Crystal structure of the 3rd PDZ domain of human discs large homologue 2, DLG2 | | 分子名称: | Discs large homolog 2 | | 著者 | Turnbull, A.P, Phillips, C, Berridge, G, Savitsky, P, Smee, C.E.A, Papagrigoriou, E, Debreczeni, J, Gorrec, F, Elkins, J.M, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
8ZPO
 
 | |
2HE4
 
 | | The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif | | 分子名称: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | | 著者 | Papagrigoriou, E, Elkins, J.M, Berridge, G, Gileady, O, Colebrook, S, Gileadi, C, Salah, E, Savitsky, P, Pantic, N, Gorrec, F, Bunkoczi, G, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Doyle, D.A, Structural Genomics Consortium (SGC) | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2RG5
 
 | | Phenylalanine pyrrolotriazine p38 alpha map kinase inhibitor compound 11B | | 分子名称: | Mitogen-activated protein kinase 14, N-ethyl-4-{[5-(methoxycarbamoyl)-2-methylphenyl]amino}-5-methylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide | | 著者 | Sack, J.S. | | 登録日 | 2007-10-02 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design, Synthesis, and Anti-inflammatory Properties of Orally Active 4-(Phenylamino)-pyrrolo[2,1-f][1,2,4]triazine p38alpha Mitogen-Activated Protein Kinase Inhibitors
J.Med.Chem., 51, 2008
|
|
2I1N
 
 | | Crystal structure of the 1st PDZ domain of Human DLG3 | | 分子名称: | Discs, large homolog 3, SODIUM ION | | 著者 | Turnbull, A.P, Phillips, C, Bunkoczi, G, Debreczeni, J, Ugochukwu, E, Pike, A.C.W, Gorrec, F, Umeano, C, Elkins, J, Berridge, G, Savitsky, P, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | 登録日 | 2006-08-14 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | 分子名称: | Granzyme M, SSGKVPL, SULFATE ION | | 著者 | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | 登録日 | 2008-01-22 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
7BZF
 
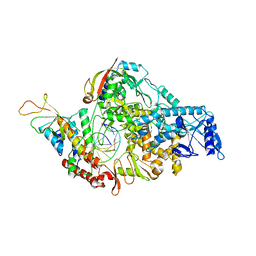 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-04-27 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
2ZGC
 
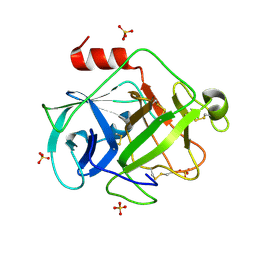 | | Crystal Structure of Active Human Granzyme M | | 分子名称: | Granzyme M, SULFATE ION | | 著者 | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | 登録日 | 2008-01-21 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
8HVR
 
 | |
4HXX
 
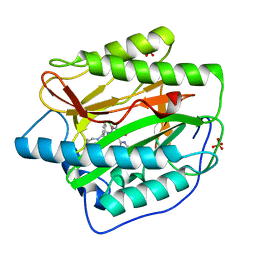 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | 分子名称: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | 登録日 | 2012-11-12 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
9C6O
 
 | |
9DAK
 
 | |
7TD5
 
 | | Structure of human PRC2-EZH1 containing phosphorylated SUZ12 | | 分子名称: | Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, Polycomb protein SUZ12, ... | | 著者 | Gong, L, Jiao, L, Liu, X. | | 登録日 | 2021-12-30 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.994 Å) | | 主引用文献 | CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site.
Nat Commun, 13, 2022
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-05-07 | | 公開日 | 2020-06-03 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
9C45
 
 | |
9C44
 
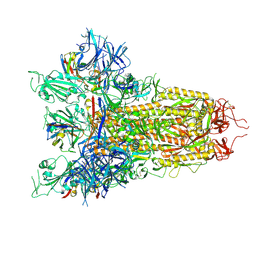 | |
7DNU
 
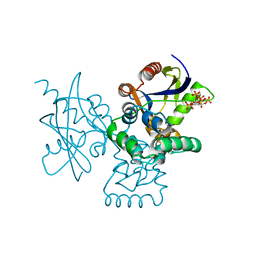 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
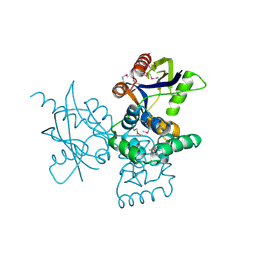 | | mRNA-decapping enzyme g5Rp | | 分子名称: | mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | 分子名称: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | 著者 | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | 登録日 | 2021-02-21 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
3BXU
 
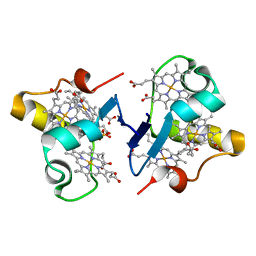 | |
8JM9
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JME
 
 | |
