1B5W
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | 分子名称: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1999-01-11 | | 公開日 | 1999-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1G0D
 
 | | CRYSTAL STRUCTURE OF RED SEA BREAM TRANSGLUTAMINASE | | 分子名称: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, SULFATE ION | | 著者 | Noguchi, K, Ishikawa, K, Yokoyama, K, Ohtsuka, T, Nio, N, Suzuki, E. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of red sea bream transglutaminase.
J.Biol.Chem., 276, 2001
|
|
2RVD
 
 | |
1B5X
 
 | | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants | | 分子名称: | PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1999-01-11 | | 公開日 | 1999-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Y
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | 分子名称: | PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1999-01-11 | | 公開日 | 1999-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5U
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | 分子名称: | PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1999-01-11 | | 公開日 | 1999-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1DJU
 
 | | CRYSTAL STRUCTURE OF AROMATIC AMINOTRANSFERASE FROM PYROCOCCUS HORIKOSHII OT3 | | 分子名称: | AROMATIC AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matsui, I, Matsui, E, Sakai, Y, Kikuchi, H, Kawarabayashi, H. | | 登録日 | 1999-12-06 | | 公開日 | 2001-04-11 | | 最終更新日 | 2018-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii.
J.Biol.Chem., 275, 2000
|
|
8IG1
 
 | |
8JYG
 
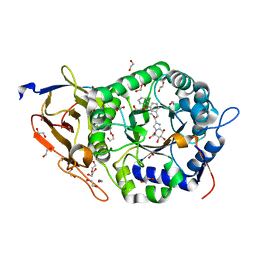 | | Crystal structure of Human HPSE1 in complex with inhibitor | | 分子名称: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mima, M, Fujimoto, N, Imai, Y. | | 登録日 | 2023-07-03 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
1J26
 
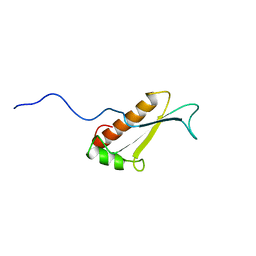 | | Solution structure of a putative peptidyl-tRNA hydrolase domain in a mouse hypothetical protein | | 分子名称: | immature colon carcinoma transcript 1 | | 著者 | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-12-25 | | 公開日 | 2004-06-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality
J.Mol.Biol., 2010
|
|
2LWI
 
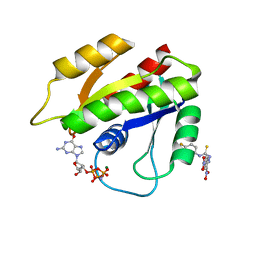 | | Solution structure of H-RasT35S mutant protein in complex with Kobe2601 | | 分子名称: | 2-(2,4-dinitrophenyl)-N-(4-fluorophenyl)hydrazinecarbothioamide, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Araki, M, Tamura, A, Shima, F, Kataoka, T. | | 登録日 | 2012-08-01 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8J6G
 
 | |
4ISU
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | 分子名称: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | 著者 | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2013-01-17 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
8WGT
 
 | | Crystal structure of V30M-TTR in complex with compound 7 | | 分子名称: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | 著者 | Yokoyama, T. | | 登録日 | 2023-09-22 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGS
 
 | | Crystal structure of V30M-TTR in complex with compound 4 | | 分子名称: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-(2-ethyl-4-iodanyl-1-benzofuran-3-yl)methanone | | 著者 | Yokoyama, T. | | 登録日 | 2023-09-22 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGU
 
 | | Crystal structure of V30M-TTR in complex with compound 20 | | 分子名称: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-[2-ethyl-4,7-bis(fluoranyl)-1-benzofuran-3-yl]methanone | | 著者 | Yokoyama, T. | | 登録日 | 2023-09-22 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.508 Å) | | 主引用文献 | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
6LVM
 
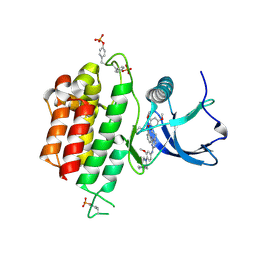 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | 分子名称: | 2-[[5-[2-(3,5-dimethoxyphenyl)ethyl]-2-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]pyrimidin-4-yl]amino]-N-ethyl-benzenesulfonamide, Fibroblast growth factor receptor 3 | | 著者 | Echizen, Y, Tateishi, Y, Amano, Y. | | 登録日 | 2020-02-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6LVK
 
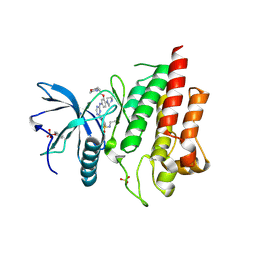 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | 分子名称: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[4-(4-methylpiperazin-1-yl)-3-(2-morpholin-4-ylethoxy)phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | 著者 | Echizen, Y, Amano, Y, Tateishi, Y. | | 登録日 | 2020-02-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
3WUS
 
 | | Crystal Structure of the Vif-Binding Domain of Human APOBEC3F | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | 著者 | Nakashima, M, Kawamura, T, Ode, H, Watanabe, N, Iwatani, Y. | | 登録日 | 2014-05-02 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Structural Insights into HIV-1 Vif-APOBEC3F Interaction.
J.Virol., 90, 2015
|
|
1JIE
 
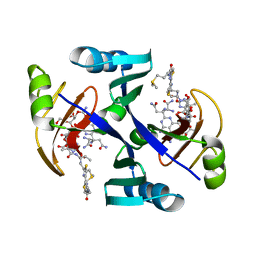 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with metal-free bleomycin | | 分子名称: | BLEOMYCIN A2, bleomycin-binding protein | | 著者 | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | 登録日 | 2001-07-02 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
7DHL
 
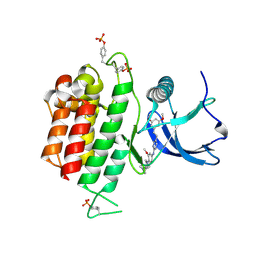 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | 分子名称: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | 著者 | Echizen, Y, Tateishi, Y, Amano, Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
1JIF
 
 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with copper(II)-bleomycin | | 分子名称: | BLEOMYCIN A2, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | 登録日 | 2001-07-02 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
6LIV
 
 | |
2TGI
 
 | |
3WO8
 
 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | 分子名称: | Beta-N-acetylglucosaminidase | | 著者 | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | 登録日 | 2013-12-20 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
