8R4X
 
 | | Structure of Chitinase-3-like protein 1 in complex with inhibitor 30 | | 分子名称: | (2~{S},5~{S})-4-[1-(4-chloranylpyridin-2-yl)piperidin-4-yl]-5-[(4-chlorophenyl)methyl]-2-methyl-morpholine, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | 登録日 | 2023-11-14 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
7Q10
 
 | |
3K1K
 
 | | Green fluorescent protein bound to enhancer nanobody | | 分子名称: | Enhancer, Green Fluorescent Protein | | 著者 | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | 登録日 | 2009-09-28 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
8R42
 
 | | Structure of CHI3L1 in complex with inhibititor 2 | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[(2~{R})-2-[(4-chlorophenyl)methyl]pyrrolidin-1-yl]piperidin-1-yl]pyridine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | 登録日 | 2023-11-10 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
5MIH
 
 | | Crystal structure of the lectin LecA from Pseudomonas aeruginosa in complex with a phenyl-epoxy-galactopyranoside | | 分子名称: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, PA-I galactophilic lectin, ... | | 著者 | Wagner, S, Hauk, D, Hofmann, M, Joachim, I, Sommer, R, Muller, R, Imberty, A, Varrot, A, Titz, A. | | 登録日 | 2016-11-28 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent Lectin Inhibition and Application in Bacterial Biofilm Imaging.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MRR
 
 | | Crystal structure of L1 protease of Lysobacter sp. XL1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of L1 protease of Lysobacter sp. XL1
To Be Published
|
|
5EQR
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with danoprevir | | 分子名称: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, SULFATE ION, ... | | 著者 | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | 登録日 | 2015-11-13 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
3K3S
 
 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | 分子名称: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | 著者 | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-10-04 | | 公開日 | 2009-10-27 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | 分子名称: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | 分子名称: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | 著者 | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-01-26 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6T28
 
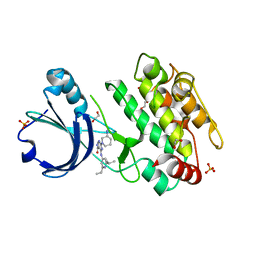 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | 分子名称: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | 著者 | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-08 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
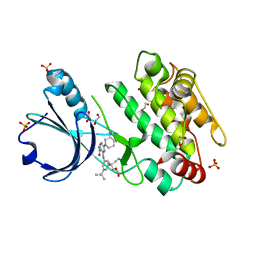 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | 分子名称: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | 著者 | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-08 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.484 Å) | | 主引用文献 | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
5HM5
 
 | |
6T36
 
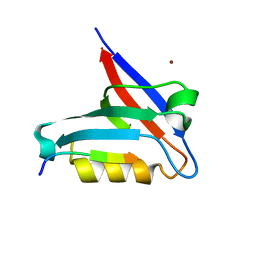 | | Crystal structure of the PTPN3 PDZ domain bound to the HBV core protein C-terminal peptide | | 分子名称: | BROMIDE ION, Capsid protein, Tyrosine-protein phosphatase non-receptor type 3 | | 著者 | Genera, M, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | 登録日 | 2019-10-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein.
Sci Rep, 11, 2021
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | 分子名称: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | 著者 | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | 登録日 | 2014-08-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3JYR
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltose-binding periplasmic protein | | 著者 | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | 登録日 | 2009-09-22 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | 分子名称: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | 著者 | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | 登録日 | 2014-08-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q1D
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, ... | | 著者 | Pontillo, N, Guskov, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | 分子名称: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
7Q1X
 
 | | Acetyltrasferase(3) type IIIa in complex with neomycin B | | 分子名称: | Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pontillo, N, Guskov, A. | | 登録日 | 2021-10-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The 3-N-alkylation of the neomycin B outmaneuvers the aminoglycoside resistant enzyme acetyltransferase(3)IIIa via an unexpected mechanism
To Be Published
|
|
7Q0Q
 
 | |
6WKP
 
 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6F8H
 
 | | antitoxin GraA | | 分子名称: | XRE family transcriptional regulator | | 著者 | Talavera, A, Loris, R. | | 登録日 | 2017-12-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
7Q1M
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2S)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | 登録日 | 2021-10-20 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | 分子名称: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | 著者 | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | 登録日 | 2020-12-08 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.873 Å) | | 主引用文献 | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
