8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | 分子名称: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | 著者 | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | 登録日 | 2023-01-31 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
5GZE
 
 | |
5GZG
 
 | |
5EOL
 
 | |
8IKH
 
 | | Cryo-EM structure of human receptor with G proteins | | 分子名称: | 3-[(1R)-1-(2-methoxyphenyl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Shen, S.Y, Shao, Z.H. | | 登録日 | 2023-02-28 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8IKG
 
 | | Cryo-EM structure of human receptor with G proteins | | 分子名称: | 3-[(1S)-1-(furan-2-yl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Shen, S.Y, Shao, Z.H. | | 登録日 | 2023-02-28 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5GQT
 
 | |
5GZC
 
 | |
8X5D
 
 | |
8J5X
 
 | | The crystal structure of TrkA(G595R) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.09192252 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J5W
 
 | | The crystal structure of TrkA(F589L) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.28041458 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
5GZD
 
 | |
5GZF
 
 | |
2JXW
 
 | |
7LIP
 
 | |
7LIN
 
 | |
7LIO
 
 | |
7LIQ
 
 | |
5AGR
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | 分子名称: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | 著者 | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | 登録日 | 2015-02-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5AGT
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | 分子名称: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | 著者 | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | 登録日 | 2015-02-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5AGS
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | 分子名称: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | 著者 | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | 登録日 | 2015-02-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5B7I
 
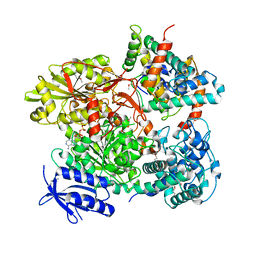 | | Cas3-AcrF3 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, ... | | 著者 | Wang, X, Zhu, Y. | | 登録日 | 2016-06-07 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3
Nat.Struct.Mol.Biol., 23, 2016
|
|
4RPV
 
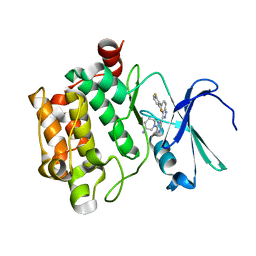 | | co-crystal structure of Pim1 with compound 3 | | 分子名称: | (3S)-1-{6-[5-(2,6-difluorophenyl)-2H-indazol-3-yl]pyrazin-2-yl}piperidin-3-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Huang, X. | | 登録日 | 2014-10-31 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | The discovery of novel 3-(pyrazin-2-yl)-1H-indazoles as potent pan-Pim kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6JK6
 
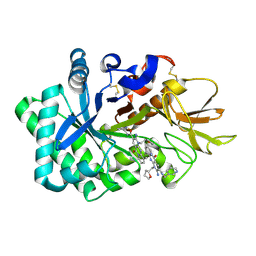 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 2-8-s2 | | 分子名称: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitotriosidase-1 | | 著者 | Jiang, X, Yang, Q. | | 登録日 | 2019-02-27 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.571 Å) | | 主引用文献 | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JKF
 
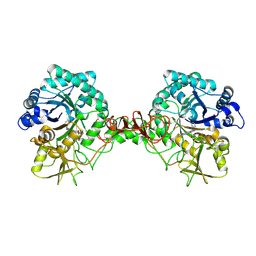 | | Crystal structure of Serratia marcescens Chitinase B complexed with compound 2-8-s2 | | 分子名称: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | 著者 | Yang, Q, Jiang, X. | | 登録日 | 2019-02-28 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
