7VQB
 
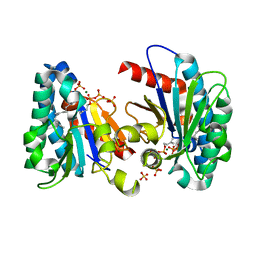 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | 分子名称: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | 著者 | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | 登録日 | 2021-10-19 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQA
 
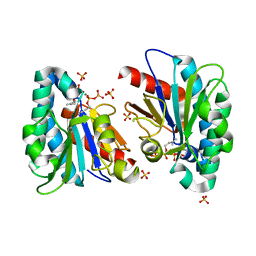 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | 分子名称: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | 著者 | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | 登録日 | 2021-10-19 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQ9
 
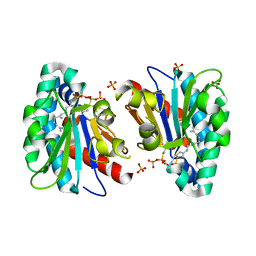 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl thiopyrophosphate and isopentyl S-thiolodiphosphate | | 分子名称: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | 著者 | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | 登録日 | 2021-10-19 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
5GWV
 
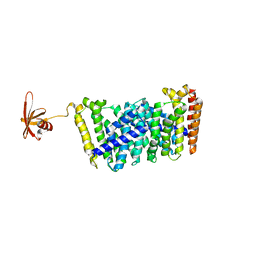 | | Structure of MoeN5-Sso7d fusion protein in complex with a substrate analogue | | 分子名称: | (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | 著者 | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | 登録日 | 2016-09-14 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5GWW
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with a permethylated substrate analogue | | 分子名称: | MoeN5,DNA-binding protein 7d, methyl (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoate | | 著者 | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | 登録日 | 2016-09-14 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8K6B
 
 | |
8K6A
 
 | |
8K68
 
 | |
8K67
 
 | |
8K6C
 
 | |
8K6D
 
 | |
8JTL
 
 | | Structure of OY phytoplasma SAP05 binding with AtRpn10 | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | 著者 | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
8JTK
 
 | | Structure of AYWB phytoplasma SAP05 recognizing AtRpn10 | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | 著者 | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | 著者 | Cao, L, Wang, X. | | 登録日 | 2021-01-18 | | 公開日 | 2021-04-21 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7CHU
 
 | | Geobacillus virus E2 - ORF18 | | 分子名称: | Putative pectin lyase | | 著者 | Gong, Y. | | 登録日 | 2020-07-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural and functional characterization of the deep-sea thermophilic bacteriophage GVE2 tailspike protein.
Int.J.Biol.Macromol., 164, 2020
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | 登録日 | 2011-12-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | 著者 | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | 登録日 | 2011-12-14 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
6LZH
 
 | | Crystal structure of Alpha/beta hydrolase GrgF from Penicillium sp. sh18 | | 分子名称: | GrgF, SODIUM ION | | 著者 | Wang, H, Yu, J, Wang, W.G, Matsuda, Y, Yao, M. | | 登録日 | 2020-02-19 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide, Gregatin A.
J.Am.Chem.Soc., 142, 2020
|
|
5Y5W
 
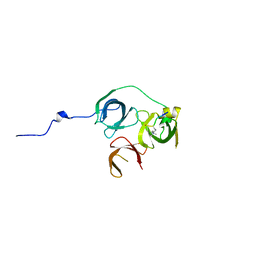 | |
4XO2
 
 | |
4GEH
 
 | |
4H1E
 
 | |
3CID
 
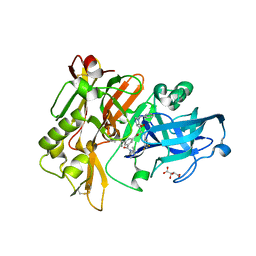 | | Structure of BACE Bound to SCH726222 | | 分子名称: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Strickland, C, Cumming, J. | | 登録日 | 2008-03-11 | | 公開日 | 2008-06-10 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIC
 
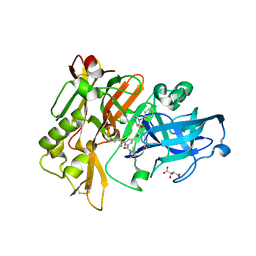 | | Structure of BACE Bound to SCH709583 | | 分子名称: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Strickland, C, Cumming, J. | | 登録日 | 2008-03-11 | | 公開日 | 2008-06-10 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2REW
 
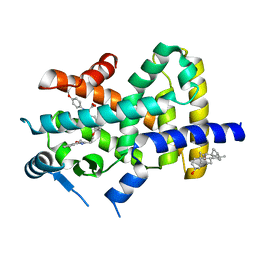 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | 分子名称: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | 著者 | Muckelbauer, J. | | 登録日 | 2007-09-27 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
