7NH7
 
 | |
7Q85
 
 | | Crystal structure of human STING in complex with MD1193 | | 分子名称: | 9-[(1R,6R,8R,13E,15R,17R,18R)-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,16-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadec-13-en-8-yl]purin-6-amine, Stimulator of interferon genes protein | | 著者 | Smola, M, Klima, M, Boura, E. | | 登録日 | 2021-11-10 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.359 Å) | | 主引用文献 | Vinylphosphonate-based cyclic dinucleotides enhance STING-mediated cancer immunotherapy.
Eur.J.Med.Chem., 259, 2023
|
|
7R1U
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | 分子名称: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
5I0N
 
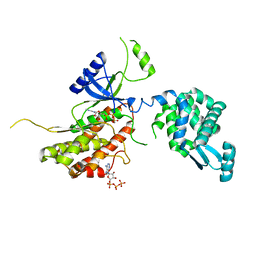 | | PI4K IIalpha bound to calcium | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Baumlova, A, Boura, E. | | 登録日 | 2016-02-04 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | PI4K IIalpha bound to calcium
To Be Published
|
|
4WAE
 
 | |
4WAG
 
 | |
7OB3
 
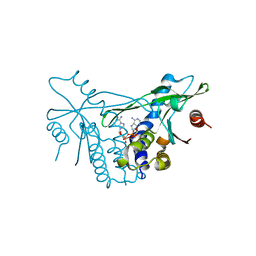 | | hSTING in complex with 3',3'-c-di-araAMP | | 分子名称: | 3',3'-c-di-araAMP, Stimulator of interferon genes protein | | 著者 | Smola, M, Boura, E. | | 登録日 | 2021-04-20 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Enzymatic Synthesis of 3'-5', 3'-5' Cyclic Dinucleotides, Their Binding Properties to the Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations
Biochemistry, 2021
|
|
4BTG
 
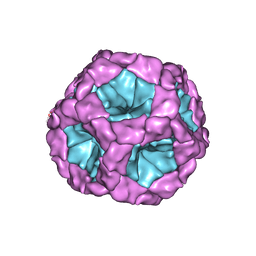 | | Coordinates of the bacteriophage phi6 capsid subunits (P1A and P1B) fitted into the cryoEM reconstruction of the procapsid at 4.4 A resolution | | 分子名称: | MAJOR INNER PROTEIN P1 | | 著者 | Nemecek, D, Boura, E, Wu, W, Cheng, N, Plevka, P, Qiao, J, Mindich, L, Heymann, J.B, Hurley, J.H, Steven, A.C. | | 登録日 | 2013-06-17 | | 公開日 | 2013-08-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Subunit Folds and Maturation Pathway of a Dsrna Virus Capsid.
Structure, 21, 2013
|
|
4BTQ
 
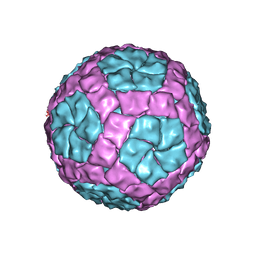 | | Coordinates of the bacteriophage phi6 capsid subunits fitted into the cryoEM map EMD-1206 | | 分子名称: | MAJOR INNER PROTEIN P1 | | 著者 | Nemecek, D, Boura, E, Wu, W, Cheng, N, Plevka, P, Qiao, J, Mindich, L, Heymann, J.B, Hurley, J.H, Steven, A.C. | | 登録日 | 2013-06-18 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Subunit Folds and Maturation Pathway of a Dsrna Virus Capsid.
Structure, 21, 2013
|
|
6FK5
 
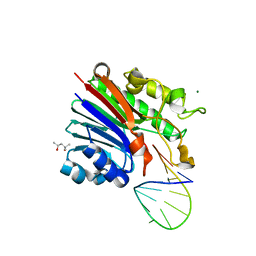 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to DNA substrate in presence of magnesium ion | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), MAGNESIUM ION, ... | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FKE
 
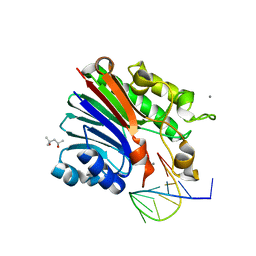 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FK4
 
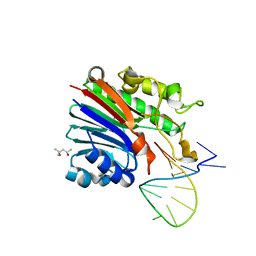 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
8BWU
 
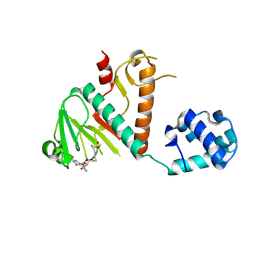 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | 著者 | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | 登録日 | 2022-12-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
8A5X
 
 | |
7ZIT
 
 | |
4WTV
 
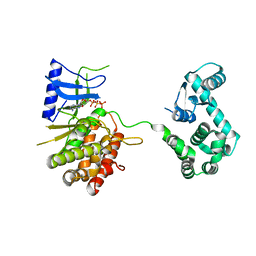 | | Crystal structure of the phosphatidylinositol 4-kinase IIbeta | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-beta,Endolysin,Phosphatidylinositol 4-kinase type 2-beta | | 著者 | Klima, M, Baumlova, A, Chalupska, D, Boura, E. | | 登録日 | 2014-10-30 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YC4
 
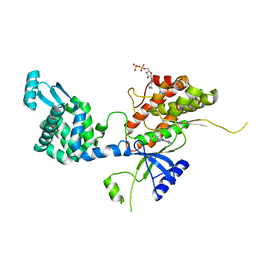 | | Crystal structure of phosphatidyl inositol 4-kinase II alpha in complex with nucleotide analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha, [(1S,3S,4S)-3-(6-amino-9H-purin-9-yl)bicyclo[2.2.1]hept-1-yl]methanol | | 著者 | Klima, M, Boura, E. | | 登録日 | 2015-02-19 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8OIV
 
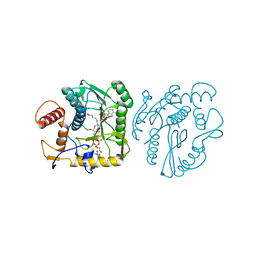 | | Monkeypox virus VP39 in complex with SAH and cap0 | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Skvara, P, Chalupska, D, Silhan, J, Klima, M, Boura, E. | | 登録日 | 2023-03-23 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural basis for RNA-cap recognition and methylation by the mpox methyltransferase VP39.
Antiviral Res., 216, 2023
|
|
8OIX
 
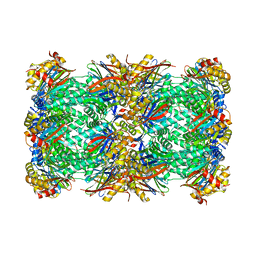 | | CryoEM structure of 20S Trichomonas vaginalis proteasome in complex with proteasome inhibitor Salinosporamid A | | 分子名称: | (3AR,6R,6AS)-6-((S)-((S)-CYCLOHEX-2-ENYL)(HYDROXY)METHYL)-6A-METHYL-4-OXO-HEXAHYDRO-2H-FURO[3,2-C]PYRROLE-6-CARBALDEHYDE, Family T1, proteasome alpha subunit, ... | | 著者 | Silhan, J, Fajtova, P, Boura, E. | | 登録日 | 2023-03-23 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Tv20S proteasome in the complex with marizomib
To Be Published
|
|
8ORV
 
 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1203 | | 分子名称: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | 著者 | Duchoslav, V, Klima, M, Boura, E. | | 登録日 | 2023-04-17 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8PEM
 
 | |
8P0T
 
 | |
8P7A
 
 | |
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | 分子名称: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | 著者 | Klima, M, Boura, E. | | 登録日 | 2023-05-19 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
