1BYF
 
 | | STRUCTURE OF TC14; A C-TYPE LECTIN FROM THE TUNICATE POLYANDROCARPA MISAKIENSIS | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Poget, S.F, Legge, G.B, Bycroft, M, Williams, R.L. | | 登録日 | 1998-10-14 | | 公開日 | 1999-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of a tunicate C-type lectin from Polyandrocarpa misakiensis complexed with D -galactose.
J.Mol.Biol., 290, 1999
|
|
5TS8
 
 | | Z. MAYS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE | | 分子名称: | 1,2-ETHANEDIOL, 5,6-DIBROMOBENZOTRIAZOLE, ACETATE ION, ... | | 著者 | Winiewska, M, Kucinska, K, Czapinska, H, Piasecka, A, Bochtler, M, Poznanski, J. | | 登録日 | 2016-10-28 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | STRUCTURAL AND THERMODYNAMIC ANALYSIS OF 5,6-DIBROMOBENZOTRIAZOLE BINDING TO CASEIN KINASE 2 ALPHA
To Be Published
|
|
1N90
 
 | |
5Y80
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | 分子名称: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | 著者 | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | 登録日 | 2017-08-18 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
7OW9
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with Cyclo-L-leucyl-L-leucine | | 分子名称: | (3S,6S)-3,6-bis(2-methylpropyl)piperazine-2,5-dione, CYPX, NITRATE ION, ... | | 著者 | Snee, M, Levy, C, Leys, D, Katariya, M, Munro, A.W. | | 登録日 | 2021-06-17 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with Cyclo-L-leucyl-L-leucine
To Be Published
|
|
6P73
 
 | | Cytochrome-C-nitrite reductase | | 分子名称: | CALCIUM ION, Cytochrome c-552, HEME C | | 著者 | Schmidt, M, Pacheco, A. | | 登録日 | 2019-06-04 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | 分子名称: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | 著者 | Sun, D, Johnson, M, Masureel, M. | | 登録日 | 2023-07-27 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | 分子名称: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | 著者 | Yang, M, Gehring, K. | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
5YPX
 
 | | Crystal structure of calaxin with magnesium | | 分子名称: | Calaxin, MAGNESIUM ION | | 著者 | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2017-11-04 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
7U97
 
 | | SAAV pH 4.0 capsid structure | | 分子名称: | Capsid protein | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2022-03-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
8DRT
 
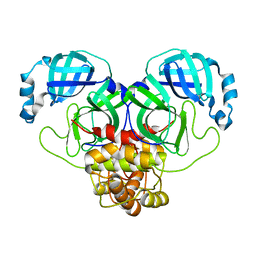 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
7U95
 
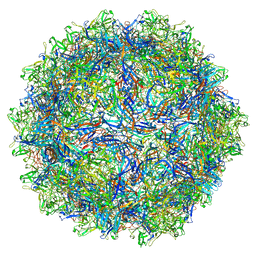 | | SAAV pH 6.0 capsid structure | | 分子名称: | Capsid protein | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2022-03-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
3FOG
 
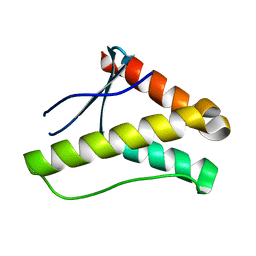 | | Crystal structure of the PX domain of sorting nexin-17 (SNX17) | | 分子名称: | SODIUM ION, Sorting nexin-17 | | 著者 | Wisniewska, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-12-30 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the PX domain of sorting nexin-17 (SNX17)
TO BE PUBLISHED
|
|
6MXT
 
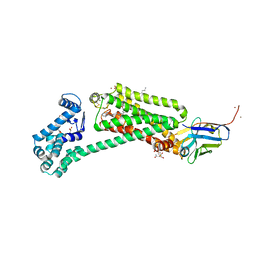 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | 著者 | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | 登録日 | 2018-10-31 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.95934224 Å) | | 主引用文献 | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6ZM7
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-EBP1 ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-01 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
6PRX
 
 | | oxidized Human Branched Chain Aminotransferase mutant C318A | | 分子名称: | Branched-chain-amino-acid aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Dong, M, Herbert, D, Gibbs, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure of an oxidized mutant of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7U96
 
 | | SAAV pH 5.5 capsid structure | | 分子名称: | Capsid protein | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2022-03-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | 分子名称: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | 著者 | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
7U94
 
 | | SAAV pH 7.4 capsid structure | | 分子名称: | Capsid protein | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2022-03-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
6P9G
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | 分子名称: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | 著者 | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-10 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8TA5
 
 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | 分子名称: | Chloride channel protein 2 | | 著者 | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | 登録日 | 2023-06-26 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
