8GYX
 
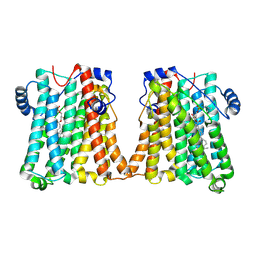 | | Cryo-EM structure of human CEPT1 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION | | 著者 | Qian, H.W, Wang, Z.H. | | 登録日 | 2022-09-24 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
8GYW
 
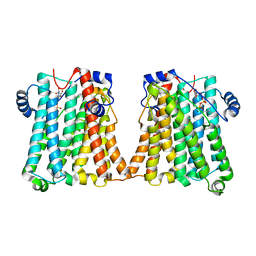 | |
8HN6
 
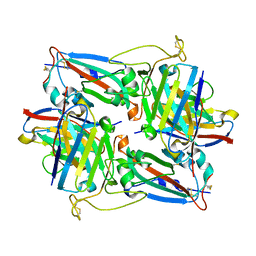 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
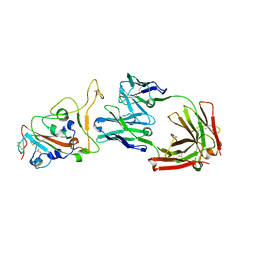 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | 分子名称: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | 登録日 | 2020-08-24 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7CUU
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | 分子名称: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | 登録日 | 2020-08-24 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
8VWP
 
 | |
7D3D
 
 | | Crystal structure of SPOP bound with a peptide | | 分子名称: | GLU-VAL-SER-ILE-ILE-GLN-GLY-ALA-ASP-SER-THR-THR, GLYCEROL, Speckle-type POZ protein | | 著者 | Yang, C.-G, Gan, J.H. | | 登録日 | 2020-09-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A peptide binder of E3 ligase adaptor SPOP disrupts oncogenic SPOP-protein interactions in kidney cancer cells.
Chin.J.Chem., 39, 2021
|
|
7UAP
 
 | |
7UAR
 
 | |
7UAQ
 
 | | Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1520 Fab Heavy Chain, ... | | 著者 | Barnes, C.O. | | 登録日 | 2022-03-13 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Immunity, 55, 2022
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | 著者 | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4WVD
 
 | | Identification of a novel FXR ligand that regulates metabolism | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Bile acid receptor, FORMIC ACID, ... | | 著者 | Wang, R, Li, Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism.
Nat Commun, 4, 2013
|
|
3KL1
 
 | |
6ORN
 
 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch bNAb 10-1074 | | 分子名称: | 10-1074 antibody Fab heavy chain, 10-1074 antibody Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | 登録日 | 2019-04-30 | | 公開日 | 2019-06-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
4WQ6
 
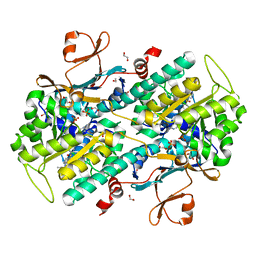 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Li, D, Wang, W. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8DD5
 
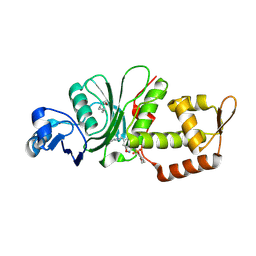 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | 分子名称: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | 著者 | Greasley, S.E, Johnson, E, Brodsky, O. | | 登録日 | 2022-06-17 | | 公開日 | 2023-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
9BFY
 
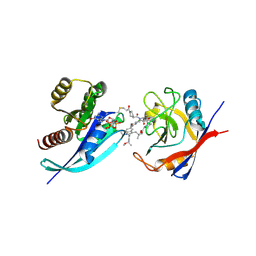 | | Tri-complex of Compound-12, KRAS G12C, and CypA | | 分子名称: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | 著者 | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | 登録日 | 2024-04-18 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Tri-complex of Compound-12, KRAS G12C, and CypA
To be published
|
|
9BG1
 
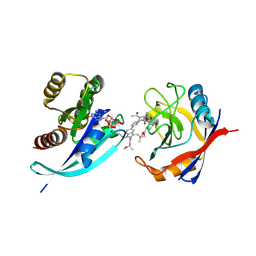 | | Tri-complex of Compound-3, KRAS G12V, and CypA | | 分子名称: | (2R)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | 著者 | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | 登録日 | 2024-04-18 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Tri-complex of Compound-3, KRAS G12V, and CypA
To be published
|
|
4LVD
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, N-(4-nitrophenyl)cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | 登録日 | 2013-07-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVB
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, N-[4-(acetylamino)phenyl]cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | 登録日 | 2013-07-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVF
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | (1S,2S)-2-phenyl-N-(pyridin-4-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | 登録日 | 2013-07-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2QKT
 
 | |
2QKU
 
 | |
2QKV
 
 | |
