2QKT
 
 | |
2QKU
 
 | |
2QKV
 
 | |
1GGX
 
 | |
2JRE
 
 | | C60-1, a PDZ domain designed using statistical coupling analysis | | Descriptor: | C60-1 PDZ domain peptide | | Authors: | Larson, C, Stiffler, M, Li, P, Rosen, M, MacBeath, G, Ranganathan, R. | | Deposit date: | 2007-06-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | C60-1, a PDZ domain designed using statistical coupling analysis
To be Published
|
|
8CTV
 
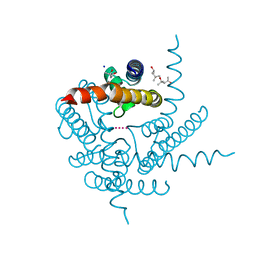 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTN
 
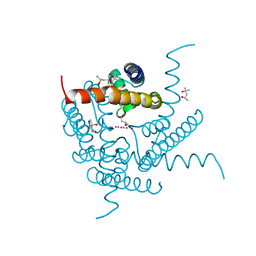 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction, no electric field) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTW
 
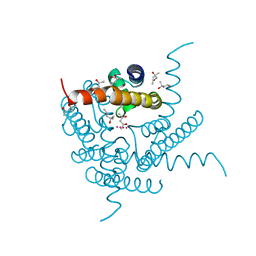 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Na+,Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTS
 
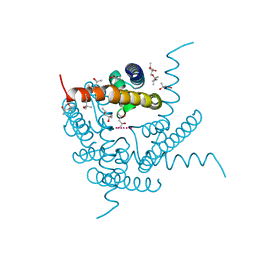 | | Room temperature crystal structure of a K+ selective NaK mutant (NaK2K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTT
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at 100K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTU
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at Room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU1
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 500ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU2
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 100ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU4
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 1us, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU3
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 200ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
5E21
 
 | | PDZ2 of LNX2 at 277K,single conformer model | | Descriptor: | Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Ranganathan, R. | | Deposit date: | 2015-09-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.011 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
5E11
 
 | | Second PDZ domain of Ligand of Numb protein X 2 by Laue crystallography (no electric field) | | Descriptor: | Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Henning, R.W, Srajer, V, Ranganathan, R. | | Deposit date: | 2015-09-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
5E22
 
 | | The second PDZ domain of Ligand of Numb protein X 2 in the presence of an electric field of ~1 MV/cm along the crystallographic x axis, with eightfold extrapolation of structure factor differences. | | Descriptor: | GLYCEROL, Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Henning, R.W, Srajer, V, Ranganathan, R. | | Deposit date: | 2015-09-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
5E1Y
 
 | | PDZ2 of LNX2 at 277K, model with alternate conformations | | Descriptor: | Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Ranganathan, R. | | Deposit date: | 2015-09-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.011 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
8CTX
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -K+,Tl+ complex | | Descriptor: | POTASSIUM ION, Potassium channel protein, THALLIUM (I) ION | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
1YMZ
 
 | | CC45, An Artificial WW Domain Designed Using Statistical Coupling Analysis | | Descriptor: | CC45 | | Authors: | Socolich, M, Lockless, S.W, Russ, W.P, Lee, H, Gardner, K.H, Ranganathan, R. | | Deposit date: | 2005-01-22 | | Release date: | 2005-09-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Evolutionary information for specifying a protein fold.
Nature, 437, 2005
|
|
1Q4B
 
 | |
1Q4E
 
 | |
1Q4A
 
 | |
1Q4D
 
 | |
