7VOC
 
 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | 分子名称: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2021-10-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.62005424 Å) | | 主引用文献 | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
 | |
8X1V
 
 | |
8HZY
 
 | |
8HZV
 
 | |
6CXD
 
 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | 分子名称: | Peptidase B, SULFATE ION | | 著者 | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-04-02 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6UUM
 
 | |
6UUL
 
 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | 著者 | Kumar, S, Wilson, I.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UUH
 
 | |
6V6W
 
 | |
7W5K
 
 | | The C296A mutant of L-sorbosone dehydrogenase (SNDH) from Gluconobacter Oxydans WSH-004 | | 分子名称: | L-sorbosone dehydrogenase, NAD(P) dependent, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | 登録日 | 2021-11-30 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7W5N
 
 | | The crystal structure of the reduced form of Gluconobacter oxydans WSH-004 SNDH | | 分子名称: | L-sorbosone dehydrogenase, NAD(P) dependent, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, D, Hou, X.D, Rao, Y.J, Yin, D.J, Zhou, J.W, Chen, J. | | 登録日 | 2021-11-30 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.988 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7W5L
 
 | | The crystal structure of the oxidized form of Gluconobacter oxydans WSH-004 SNDH | | 分子名称: | L-sorbosone dehydrogenase, NAD(P) dependent | | 著者 | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | 登録日 | 2021-11-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.980013 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | 著者 | Zhou, J.H, Hou, X.L. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.899313 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02027535 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | 分子名称: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | 著者 | Hong, W, Chen, H, Wu, Q, Lin, T. | | 登録日 | 2019-09-23 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.45 Å) | | 主引用文献 | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-07 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
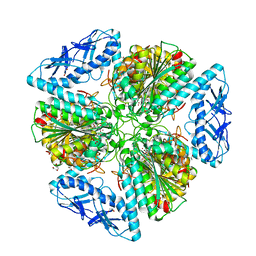 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | 分子名称: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-03-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
4Y87
 
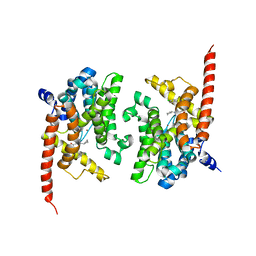 | | Crystal structure of phosphodiesterase 9 in complex with (R)-C33 (6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) | | 分子名称: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4Y8C
 
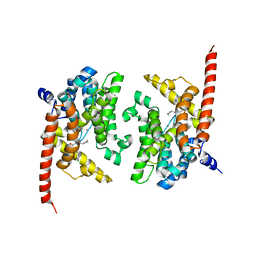 | | Crystal structure of phosphodiesterase 9 in complex with (S)-C33 | | 分子名称: | 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4Y86
 
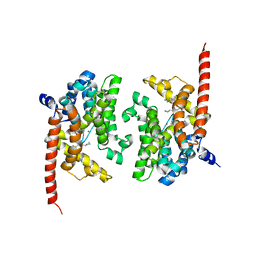 | | Crystal structure of PDE9 in complex with racemic inhibitor C33 | | 分子名称: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | 分子名称: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
6LCG
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | 分子名称: | DI(HYDROXYETHYL)ETHER, N-carbamoyl-D-amino-acid hydrolase | | 著者 | Liu, Y.F, Ni, Y, Xu, G.C, Dai, W. | | 登録日 | 2019-11-18 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
6LEI
 
 | |
