2OO8
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | 分子名称: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | 著者 | Bellon, S.F, Kim, J. | | 登録日 | 2007-01-25 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | 分子名称: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | 著者 | Noeske, J, Shu, W, Bellamacina, C. | | 登録日 | 2021-01-26 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
2OSC
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | 分子名称: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | 著者 | Bellon, S.F, Kim, J. | | 登録日 | 2007-02-05 | | 公開日 | 2007-03-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5UIX
 
 | |
4NFW
 
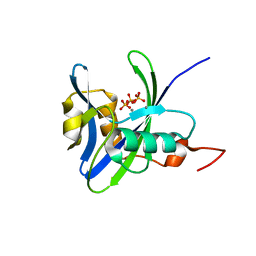 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | 分子名称: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | 著者 | Hong, M.K, Kim, J.K, Kang, L.W. | | 登録日 | 2013-11-01 | | 公開日 | 2014-05-14 | | 最終更新日 | 2015-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | 分子名称: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Poulos, T.L, Kim, J, Murarka, V.C. | | 登録日 | 2021-08-31 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.40000689 Å) | | 主引用文献 | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
4NFX
 
 | |
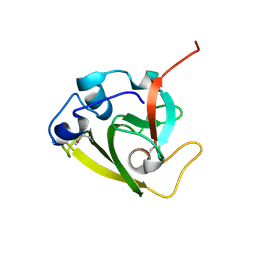
6WIN
 
 | |
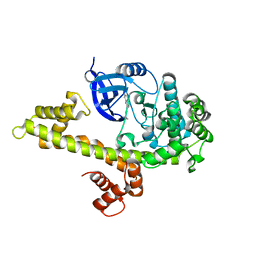
4M84
 
 | |
2KI2
 
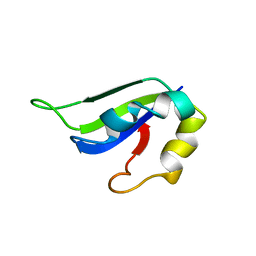 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | 分子名称: | Ss-DNA binding protein 12RNP2 | | 著者 | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | 登録日 | 2009-04-20 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
6JN8
 
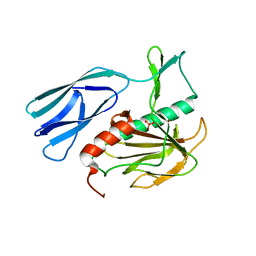 | | Structure of H216A mutant open form peptidoglycan peptidase | | 分子名称: | Peptidase M23, SULFATE ION, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
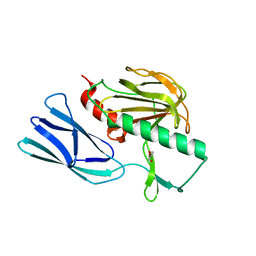 | | Structure of open form of peptidoglycan peptidase | | 分子名称: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
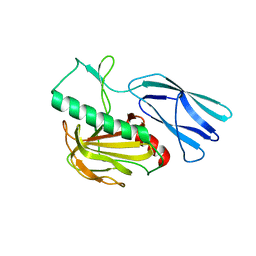 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | 分子名称: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
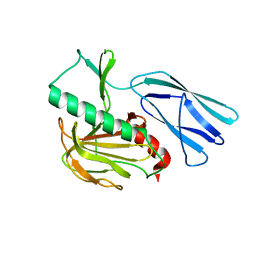 | | Structure of H247A mutant open form peptidoglycan peptidase | | 分子名称: | Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | 分子名称: | C0O-DAL-API, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.164 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | 分子名称: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
 | | Structure of wild type closed form of peptidoglycan peptidase | | 分子名称: | CITRIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5V7R
 
 | |
5V7U
 
 | |
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | 著者 | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | 登録日 | 2014-05-19 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
6KV1
 
 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | 分子名称: | CITRIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-09-03 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.722 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
7CTP
 
 | |
8EJO
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
5VBA
 
 | |
