7RLL
 
 | | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate | | 分子名称: | Arf3p, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Stogios, P.J, Michalska, K, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-25 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate
To Be Published
|
|
5UXD
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | 分子名称: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | 著者 | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | 分子名称: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | 著者 | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.794 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXC
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | 分子名称: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-08-16 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
3TYK
 
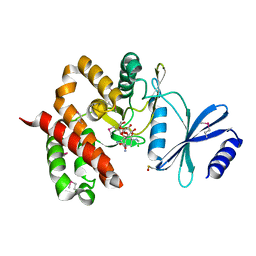 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | 分子名称: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | 著者 | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-26 | | 公開日 | 2011-10-12 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
1N2X
 
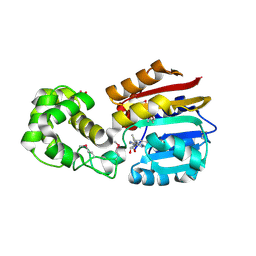 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAM | | 分子名称: | S-ADENOSYLMETHIONINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | 著者 | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-10-24 | | 公開日 | 2003-01-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
7JM1
 
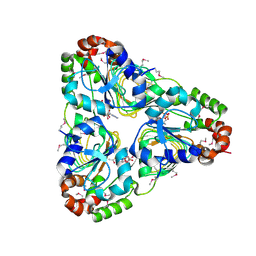 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
7JM2
 
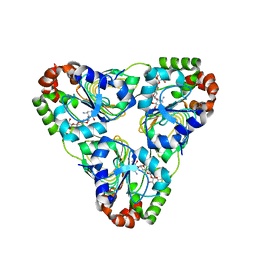 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | 分子名称: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
7JM0
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme | | 分子名称: | Aminocyclitol acetyltransferase ApmA, SULFATE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme
To Be Published
|
|
7SQO
 
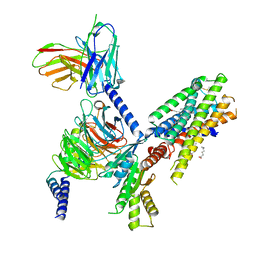 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | McGrath, A.P, Kang, Y, Flinspach, M. | | 登録日 | 2021-11-05 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
7SR8
 
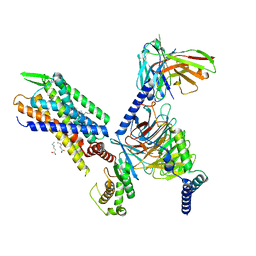 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | 著者 | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | 登録日 | 2021-11-08 | | 公開日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
7U5T
 
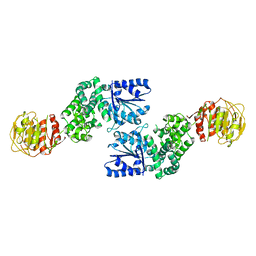 | |
7U5U
 
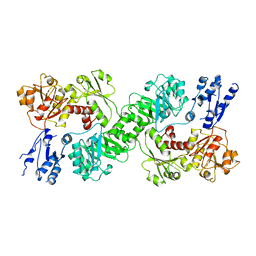 | |
7U5S
 
 | |
1KS2
 
 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | 分子名称: | protein EC1268, RPIA | | 著者 | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-01-10 | | 公開日 | 2002-08-14 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | 分子名称: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | 著者 | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2018-08-14 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
1X8H
 
 | |
1M6Y
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAH | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | 著者 | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-07-17 | | 公開日 | 2003-01-28 | | 最終更新日 | 2016-03-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
5D9M
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG | | 分子名称: | B-1,4-endoglucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9P
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG | | 分子名称: | B-1,4-endoglucanase, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-N-acetyl-beta-D-glucopyranosylamine | | 著者 | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9N
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG | | 分子名称: | B-1,4-endoglucanase, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9O
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose | | 分子名称: | B-1,4-endoglucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
1NYN
 
 | | Solution NMR Structure of Protein YHR087W from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YTYST425. | | 分子名称: | Hypothetical 12.0 kDa protein in NAM8-GAR1 intergenic region | | 著者 | Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-02-13 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | 分子名称: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | 著者 | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | 登録日 | 2022-09-10 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
3F7Z
 
 | | X-ray Co-Crystal Structure of Glycogen Synthase Kinase 3beta in Complex with an Inhibitor | | 分子名称: | 2-(1,3-benzodioxol-5-yl)-5-[(3-fluoro-4-methoxybenzyl)sulfanyl]-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | 著者 | Mol, C.D, Dougan, D.R. | | 登録日 | 2008-11-10 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
