2DUU
 
 | | Crystal Structure of apo-form of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. | | 分子名称: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | 著者 | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | 登録日 | 2006-07-27 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of apo-glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2C78
 
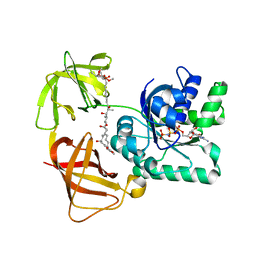 | | EF-Tu complexed with a GTP analog and the antibiotic pulvomycin | | 分子名称: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, ELONGATION FACTOR TU-A, MAGNESIUM ION, ... | | 著者 | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | 登録日 | 2005-11-18 | | 公開日 | 2006-03-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
2C77
 
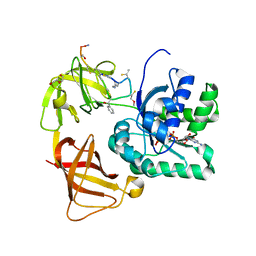 | | EF-Tu complexed with a GTP analog and the antibiotic GE2270 A | | 分子名称: | DI(HYDROXYETHYL)ETHER, ELONGATION FACTOR TU-B, MAGNESIUM ION, ... | | 著者 | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | 登録日 | 2005-11-18 | | 公開日 | 2006-03-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
2DD4
 
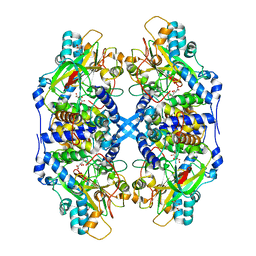 | | Thiocyanate hydrolase (SCNase) from Thiobacillus thioparus recombinant apo-enzyme | | 分子名称: | L(+)-TARTARIC ACID, Thiocyanate hydrolase alpha subunit, Thiocyanate hydrolase beta subunit, ... | | 著者 | Arakawa, T, Kawano, Y, Kataoka, S, Katayama, Y, Kamiya, N, Yohda, M, Odaka, M. | | 登録日 | 2006-01-19 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center.
J.Mol.Biol., 366, 2007
|
|
2DD5
 
 | | Thiocyanate hydrolase (SCNase) from Thiobacillus thioparus native holo-enzyme | | 分子名称: | COBALT (III) ION, SULFATE ION, Thiocyanate hydrolase alpha subunit, ... | | 著者 | Arakawa, T, Kawano, Y, Kataoka, S, Katayama, Y, Kamiya, N, Yohda, M, Odaka, M. | | 登録日 | 2006-01-19 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center.
J.Mol.Biol., 366, 2007
|
|
3WMW
 
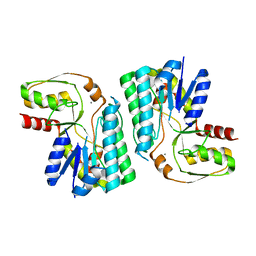 | | GalE-like L-Threonine dehydrogenase from Cupriavidus necator (apo form) | | 分子名称: | CALCIUM ION, NAD dependent epimerase/dehydratase | | 著者 | Nakano, S, Okazaki, S, Tokiwa, H, Asano, Y. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Binding of NAD+ and L-Threonine Induces Stepwise Structural and Flexibility Changes in Cupriavidus necator L-Threonine Dehydrogenase
J.Biol.Chem., 289, 2014
|
|
3WMX
 
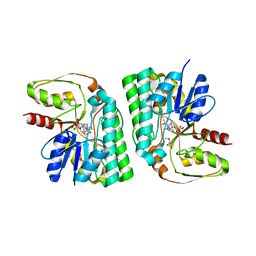 | | GalE-like L-Threonine dehydrogenase from Cupriavidus necator (holo form) | | 分子名称: | NAD dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THREONINE | | 著者 | Nakano, S, Okazaki, S, Tokiwa, H, Asano, Y. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Binding of NAD+ and L-Threonine Induces Stepwise Structural and Flexibility Changes in Cupriavidus necator L-Threonine Dehydrogenase
J.Biol.Chem., 289, 2014
|
|
3X3H
 
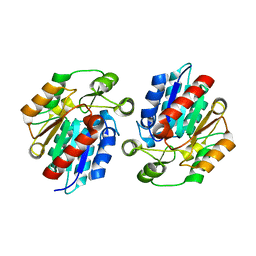 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | 分子名称: | (S)-hydroxynitrile lyase | | 著者 | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | 登録日 | 2015-01-21 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
4WUA
 
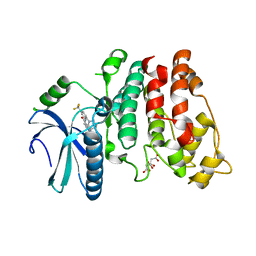 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | 分子名称: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | 著者 | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | 登録日 | 2014-10-31 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
4DG1
 
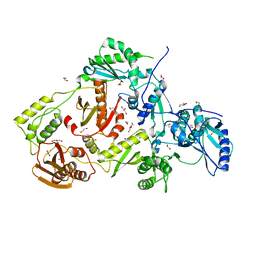 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | 登録日 | 2012-01-24 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
4MJI
 
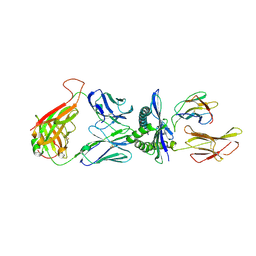 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | 分子名称: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | 登録日 | 2013-09-03 | | 公開日 | 2014-05-28 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
2Z6D
 
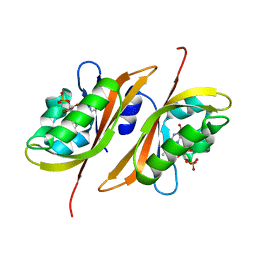 | |
2Z6C
 
 | |
5HGH
 
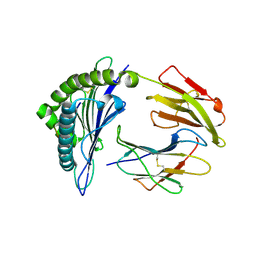 | | HLA*A2402 complexed with HIV nef138 10mer epitope | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGB
 
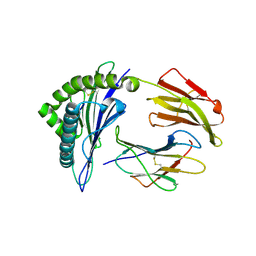 | | HLA*A2402 complexed with HIV nef138 8mer epitope | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGD
 
 | | HLA*A2402 complexed with HIV nef138 Y2F mutant 10mer epitope | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGA
 
 | | HLA*A2402 complex with HIV nef138 Y2F-8mer mutant epitope | | 分子名称: | 8-mer from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
2FMY
 
 | |
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | 著者 | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | 登録日 | 2021-07-17 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | 著者 | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | 登録日 | 2021-07-17 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | 著者 | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | 登録日 | 2021-07-17 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6L3R
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | 分子名称: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Miyahara, S, Chong, K.T, Suzuki, T. | | 登録日 | 2019-10-15 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
4YT0
 
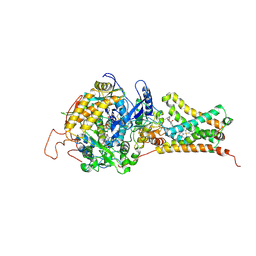 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide. | | 分子名称: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | 登録日 | 2015-03-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.66 Å) | | 主引用文献 | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSY
 
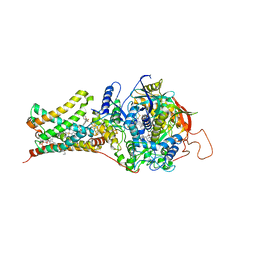 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[(2,4-dichlorophenyl)methyl]-2-(trifluoromethyl)benzamide | | 分子名称: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | 登録日 | 2015-03-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSX
 
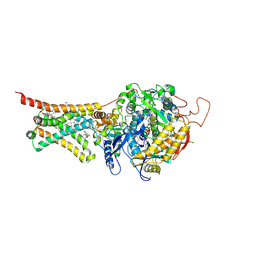 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with the specific inhibitor NN23 | | 分子名称: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | 登録日 | 2015-03-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
