3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | 分子名称: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
8T5M
 
 | | SOS2 crystal structure with fragment bound (compound 14) | | 分子名称: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-14 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5G
 
 | | SOS2 co-crystal structure with fragment bound (compound 12) | | 分子名称: | DIMETHYL SULFOXIDE, SULFATE ION, Son of sevenless homolog 2, ... | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-13 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5R
 
 | | SOS2 crystal structure with fragment bound (compound 13) | | 分子名称: | 4-(aminomethyl)benzene-1-sulfonamide, SULFATE ION, Son of sevenless homolog 2 | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-14 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
7SH0
 
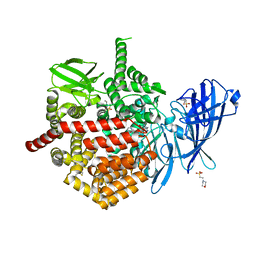 | | CRYSTAL STRUCTURE OF ENDOPLASMIC RETICULUM AMINOPEPTIDASE 2 (ERAP2) COMPLEX WITH A HIGHLY SELECTIVE AND POTENT SMALL MOLECULE | | 分子名称: | (2S)-N-hydroxy-3-(4-methoxyphenyl)-2-[4-({[5-(pyridin-2-yl)thiophene-2-sulfonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Li, L, Bouvier, M. | | 登録日 | 2021-10-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6AVH
 
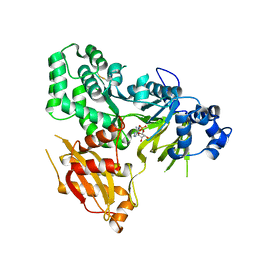 | | GH3.15 acyl acid amido synthetase | | 分子名称: | ADENOSINE MONOPHOSPHATE, GH3.15 acyl acid amido synthetase | | 著者 | Sherp, A.M, Jez, J.M. | | 登録日 | 2017-09-02 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.011 Å) | | 主引用文献 | Arabidopsis thalianaGH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid.
J. Biol. Chem., 293, 2018
|
|
7UNJ
 
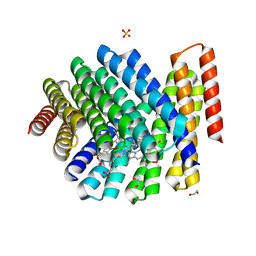 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
8UF2
 
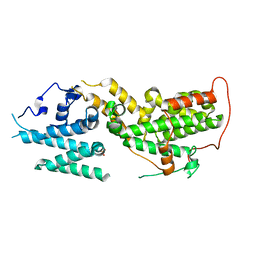 | |
8UC9
 
 | |
8U4W
 
 | |
8UH0
 
 | |
7UKS
 
 | | Crystal structure of SOS1 with phthalazine inhibitor bound (compound 15) | | 分子名称: | 4-methyl-N-{(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}-7-(piperazin-1-yl)phthalazin-1-amine, Son of sevenless homolog 1 | | 著者 | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | 登録日 | 2022-04-01 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7UKR
 
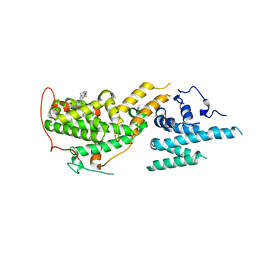 | | Crystal Structure of SOS1 with MRTX0902, a Potent and Selective Inhibitor of the SOS1:KRAS Protein-Protein Interaction | | 分子名称: | 2-methyl-3-[(1R)-1-{[4-methyl-7-(morpholin-4-yl)pyrido[3,4-d]pyridazin-1-yl]amino}ethyl]benzonitrile, Son of sevenless homolog 1 | | 著者 | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | 登録日 | 2022-04-01 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7LEZ
 
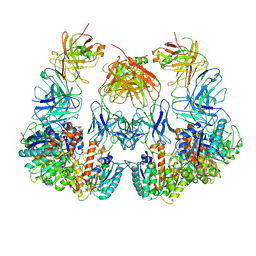 | | Trimeric human Arginase 1 in complex with mAb1 - 2 hArg:2 mAb1 complex | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb1 heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEY
 
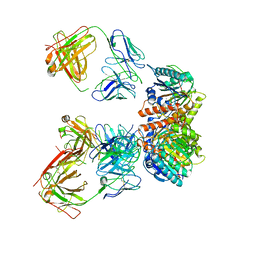 | | Trimeric human Arginase 1 in complex with mAb5 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEX
 
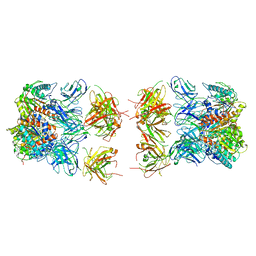 | |
7LF0
 
 | |
7LF2
 
 | | Trimeric human Arginase 1 in complex with mAb4 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF1
 
 | | Trimeric human Arginase 1 in complex with mAb3 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
3I4Q
 
 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | 分子名称: | APC40078, SODIUM ION | | 著者 | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-07-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6R2G
 
 | | Crystal structure of a single-chain protein mimetic of the gp41 NHR trimer in complex with the synthetic CHR peptide C34 | | 分子名称: | Envelope glycoprotein gp160, PHOSPHATE ION, Single-chain protein mimetics of the N-terminal heptad-repeat region of gp41 | | 著者 | Camara-Artigas, A, Conejero-Lara, F, Jurado, S, Cano-Munoz, M, Morel, B. | | 登録日 | 2019-03-17 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Thermodynamic Analysis of HIV-1 Fusion Inhibition Using Small gp41 Mimetic Proteins.
J.Mol.Biol., 431, 2019
|
|
3IRU
 
 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | 分子名称: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | 著者 | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
7QQ3
 
 | | Cryo-EM structure of the E.coli 50S ribosomal subunit in complex with the antibiotic Myxovalargin A. | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Koller, T.O, Beckert, B, Wilson, D.N. | | 登録日 | 2022-01-06 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
