8WAR
 
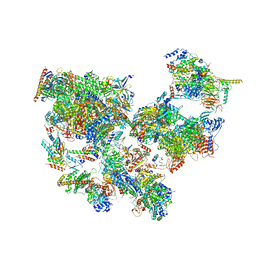 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | 分子名称: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | 著者 | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8IUE
 
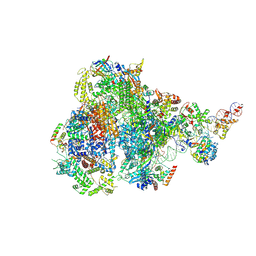 | | RNA polymerase III pre-initiation complex melting complex 1 | | 分子名称: | DNA (74-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2023-03-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8IUH
 
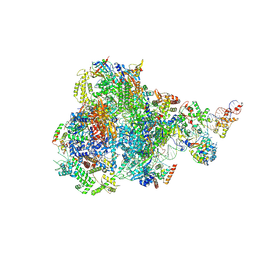 | | RNA polymerase III pre-initiation complex open complex 1 | | 分子名称: | DNA (81-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2023-03-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8ITY
 
 | | human RNA polymerase III pre-initiation complex closed DNA 1 | | 分子名称: | DNA (82-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2023-03-23 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | 著者 | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | 登録日 | 2016-12-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
8W8E
 
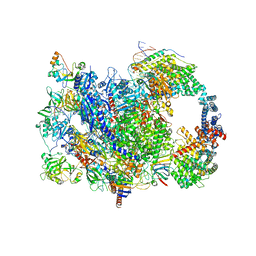 | | human co-transcriptional RNA capping enzyme RNGTT | | 分子名称: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | 著者 | Li, Y, Wang, Q, Xu, Y, Li, Z. | | 登録日 | 2023-09-02 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W8F
 
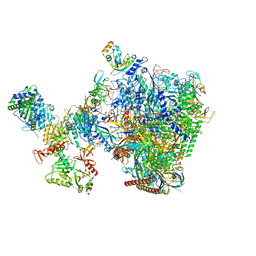 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | 分子名称: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | 著者 | Li, Y, Wang, Q, Xu, Y, Li, Z. | | 登録日 | 2023-09-02 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | 分子名称: | Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5VMC
 
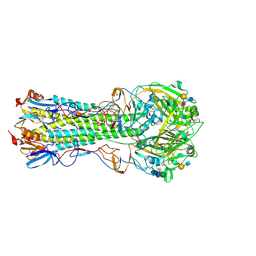 | | Influenza hemagglutinin H1 mutant DH1 in complex with 6'SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Ni, F, Kondrashkina, E, Wang, Q. | | 登録日 | 2017-04-27 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Determinant of receptor-preference switch in influenza hemagglutinin.
Virology, 513, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
8HIT
 
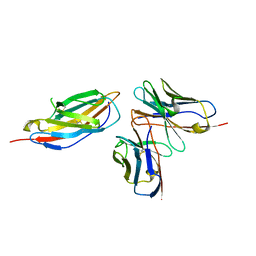 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | 分子名称: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | 著者 | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | 登録日 | 2022-11-21 | | 公開日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | 分子名称: | Major prion protein | | 著者 | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-01-18 | | 公開日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | 著者 | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | 登録日 | 2020-09-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | 登録日 | 2020-09-02 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
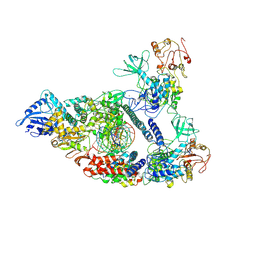 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | 登録日 | 2020-09-02 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7D6V
 
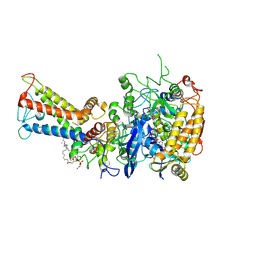 | | Mycobacterium smegmatis Sdh1 in complex with UQ1 | | 分子名称: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Zhou, X, Gao, Y, Wang, Q, Gong, H, Rao, Z. | | 登録日 | 2020-10-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D6X
 
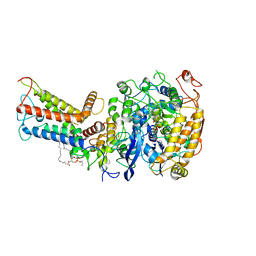 | | Mycobacterium smegmatis Sdh1 complex in the apo form | | 分子名称: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Zhou, X, Gao, Y, Wang, Q, Gong, H, Rao, Z. | | 登録日 | 2020-10-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V4Y
 
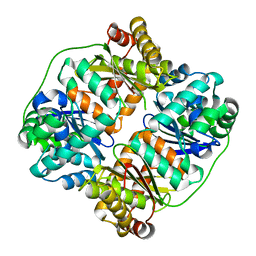 | | TTHA1264/TTHA1265 complex | | 分子名称: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | 著者 | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | 登録日 | 2021-08-16 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | 分子名称: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | 著者 | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | 登録日 | 2020-07-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7CZB
 
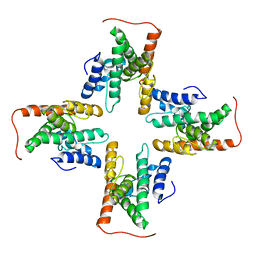 | | The cryo-EM structure of the ERAD retrotranslocation channel formed by human Derlin-1 | | 分子名称: | Derlin-1 | | 著者 | Rao, B, Li, S, Yao, D, Wang, Q, Xia, Y, Jia, Y, Shen, Y, Cao, Y. | | 登録日 | 2020-09-07 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1.
Sci Adv, 7, 2021
|
|
7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | 著者 | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | 登録日 | 2020-08-06 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
7VZF
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-11-16 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
