4W9I
 
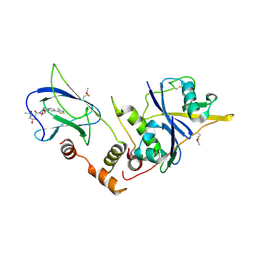 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((2S,4R)-1-acetyl-4-hydroxypyrrolidine-2-carbonyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 10) | | 分子名称: | (4R)-1-acetyl-4-hydroxy-L-prolyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | 著者 | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | 登録日 | 2014-08-27 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9T
 
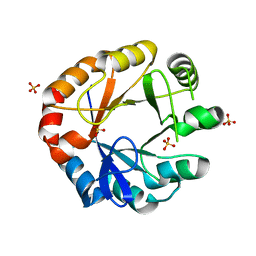 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | 分子名称: | Phosphoribosyl isomerase A, SULFATE ION | | 著者 | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-27 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4V0I
 
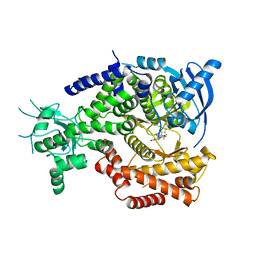 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | 分子名称: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | 著者 | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | 登録日 | 2014-09-16 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4USP
 
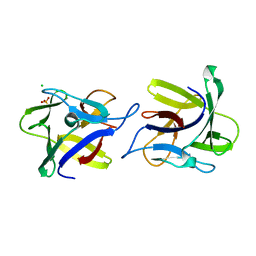 | | X-ray structure of the dimeric CCL2 lectin in native form | | 分子名称: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | 著者 | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | 登録日 | 2014-07-11 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4UWE
 
 | |
4V2A
 
 | | human Unc5A ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | 著者 | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | 登録日 | 2014-10-08 | | 公開日 | 2014-11-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4V42
 
 | | Crystal structure of the ribosome at 5.5 A resolution. | | 分子名称: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Yusupov, M.M, Yusupova, G.Z, Baucom, A, Lieberman, K, Earnest, T.N, Cate, J.H.D, Noller, H.F. | | 登録日 | 2001-03-30 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Crystal structure of the ribosome at 5.5 A resolution
Science, 292, 2001
|
|
4V7P
 
 | | Recognition of the amber stop codon by release factor RF1. | | 分子名称: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Korostelev, A, Zhu, J, Asahara, H, Noller, H.F. | | 登録日 | 2010-04-29 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.62 Å) | | 主引用文献 | Recognition of the amber UAG stop codon by release factor RF1.
Embo J., 29, 2010
|
|
4V88
 
 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | 分子名称: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | 著者 | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | 登録日 | 2011-10-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
5I6S
 
 | | Crystal structure of an endoglucanase from Penicillium verruculosum | | 分子名称: | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, endoglucanase | | 著者 | Nemashklaov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinitsyn, A. | | 登録日 | 2016-02-16 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of an endoglucanase from Penicillium verruculosum
To Be Published
|
|
4V4H
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | 分子名称: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | 著者 | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | 登録日 | 2006-08-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4W5C
 
 | | Crystal structure analysis of cruzain with three Fragments: 1 (N-(1H-benzimidazol-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide), 6 (2-amino-4,6-difluorobenzothiazole) and 9 (N-(1H-benzimidazol-2-yl)-3-(4-fluorophenyl)-1H-pyrazole-4-carboxamide). | | 分子名称: | 4,6-difluoro-1,3-benzothiazol-2-amine, Cruzipain, N-(1H-benzimidazol-2-yl)-1,3-dimethyl-1H-pyrazole-4-carboxamide, ... | | 著者 | Tochowicz, A, McKerrow, J.H, Craik, C.S. | | 登録日 | 2014-08-17 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Applying Fragments Based- Drug Design to identify multiple binding modes on cysteine protease.
To Be Published
|
|
4V4E
 
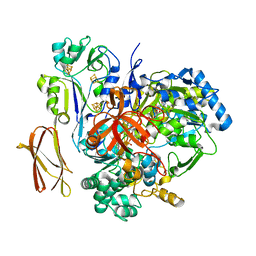 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | 著者 | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | 登録日 | 2004-06-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | 登録日 | 2007-06-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | 分子名称: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | 著者 | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | 登録日 | 2010-11-26 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.93 Å) | | 主引用文献 | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V52
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with neomycin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | 登録日 | 2007-06-15 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V9D
 
 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | 登録日 | 2012-07-31 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
4V63
 
 | | Structural basis for translation termination on the 70S ribosome. | | 分子名称: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | 著者 | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | 登録日 | 2008-05-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.207 Å) | | 主引用文献 | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
8V15
 
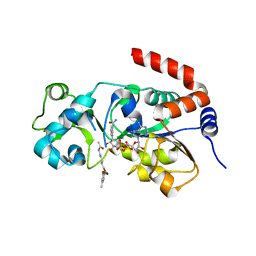 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | 分子名称: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-19 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V2N
 
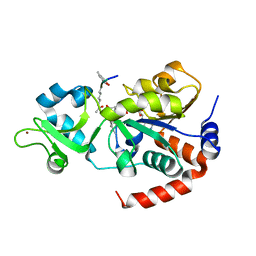 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | 分子名称: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-23 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | 分子名称: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | 著者 | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | 登録日 | 2007-07-18 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.83 Å) | | 主引用文献 | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V5D
 
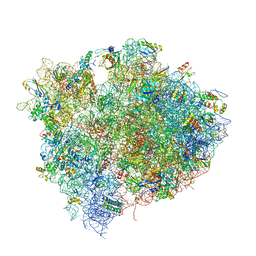 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | 分子名称: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2009-03-24 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5T
 
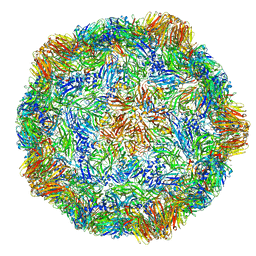 | | X-ray structure of the Grapevine Fanleaf virus | | 分子名称: | COAT PROTEIN | | 著者 | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | 登録日 | 2011-02-01 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
4V9R
 
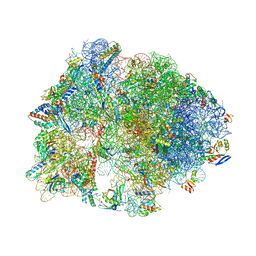 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4XCT
 
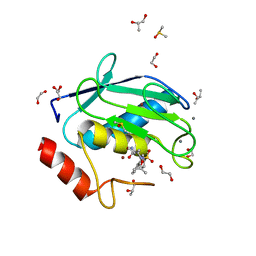 | | Crystal structure of a hydroxamate based inhibitor ARP101 (EN73) in complex with the MMP-9 catalytic domain. | | 分子名称: | (2S,3S)-butane-2,3-diol, (2~{R})-3-methyl-~{N}-oxidanylidene-2-[(4-phenylphenyl)sulfonyl-propan-2-yloxy-amino]butanamide, 1,2-ETHANEDIOL, ... | | 著者 | Stura, E.A, Tepshi, L, Nuti, E, Dive, V, Cassar-Lajeunesse, E, Vera, L, Rossello, A. | | 登録日 | 2014-12-18 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
