5VE8
 
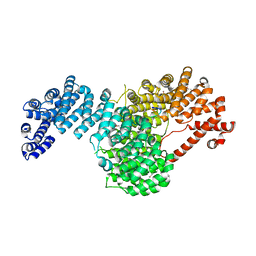 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H3 1-28 | | 分子名称: | Histone H3, Kap123 | | 著者 | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | 登録日 | 2017-04-04 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
4QBR
 
 | |
6TL8
 
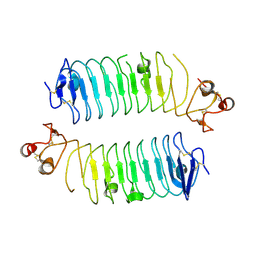 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | 著者 | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | 登録日 | 2019-12-02 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
6VAT
 
 | |
6VC7
 
 | |
6VDF
 
 | |
3H0T
 
 | | Hepcidin-Fab complex | | 分子名称: | Fab fragment, Heavy chain, Light chain, ... | | 著者 | Syed, R, Li, V. | | 登録日 | 2009-04-10 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2K21
 
 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | 分子名称: | Potassium voltage-gated channel subfamily E member | | 著者 | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | 登録日 | 2008-03-19 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | 分子名称: | Diacylglycerol kinase | | 著者 | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | 登録日 | 2009-01-06 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
2LYQ
 
 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
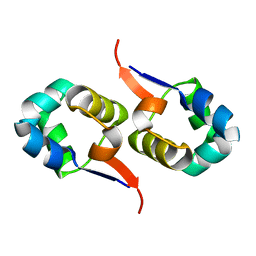 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
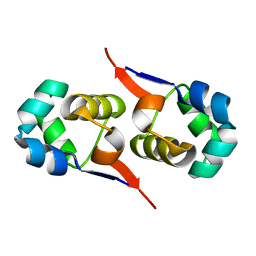 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
6I60
 
 | |
3SJF
 
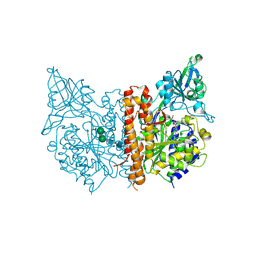 | | X-ray structure of human glutamate carboxypeptidase II in complex with a urea-based inhibitor (A25) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | 登録日 | 2011-06-21 | | 公開日 | 2011-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJG
 
 | | Human glutamate carboxypeptidase II (E424A inactive mutant ) in complex with N-acetyl-aspartyl-aminooctanoic acid | | 分子名称: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | 登録日 | 2011-06-21 | | 公開日 | 2011-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJE
 
 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-aminononanoic acid | | 分子名称: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | 登録日 | 2011-06-21 | | 公開日 | 2011-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJX
 
 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-methionine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | 登録日 | 2011-06-22 | | 公開日 | 2011-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
2IJN
 
 | |
5Z9G
 
 | | Crystal structure of KAI2 | | 分子名称: | Probable esterase KAI2 | | 著者 | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | 登録日 | 2018-02-03 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
8JZM
 
 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | 分子名称: | Toll/interleukin-1 receptor domain-containing adapter protein | | 著者 | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | 登録日 | 2023-07-05 | | 公開日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
7T63
 
 | |
