6XJZ
 
 | |
6XJW
 
 | |
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | 分子名称: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | 著者 | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | 分子名称: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | 著者 | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | 登録日 | 2011-12-14 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (8.2 Å) | | 主引用文献 | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
4TNK
 
 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (5.2 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6YL4
 
 | | Soluble epoxide hydrolase in complex with 3-((R)-3-(1-hydroxyureido)but-1-yn-1-yl)-N-((S)-3-phenyl-3-(4-trifluoromethoxy)phenyl)propyl)benzamide | | 分子名称: | 3-[(3~{R})-3-[aminocarbonyl(oxidanyl)amino]but-1-ynyl]-~{N}-[(3~{S})-3-phenyl-3-[4-(trifluoromethyloxy)phenyl]propyl]benzamide, Bifunctional epoxide hydrolase 2 | | 著者 | Kramer, J.S, Pogoryelov, D, Hiesinger, K, Proschak, E. | | 登録日 | 2020-04-06 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Design, Synthesis, and Structure-Activity Relationship Studies of Dual Inhibitors of Soluble Epoxide Hydrolase and 5-Lipoxygenase.
J.Med.Chem., 63, 2020
|
|
9DI6
 
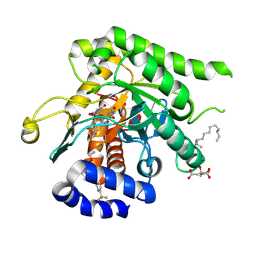 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | | 分子名称: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, CITRIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Tomchick, D, Phillips, M. | | 登録日 | 2024-09-05 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKY
 
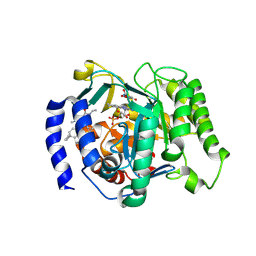 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1174 (3-((7-azaspiro[3.5]nonan-7-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | 分子名称: | 3-[(7-azaspiro[3.5]nonan-7-yl)methyl]-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Tomchick, D, Phillips, M. | | 登録日 | 2024-09-10 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1211 ((R)-3-(amino(6-(trifluoromethyl)pyridin-3-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | 分子名称: | 3-{(R)-amino[6-(trifluoromethyl)pyridin-3-yl]methyl}-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Tomchic, D, Phillips, M. | | 登録日 | 2024-09-11 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLK
 
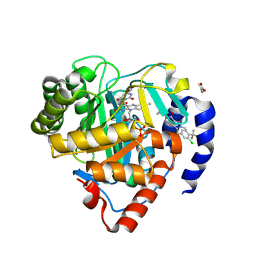 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1398 ((S)-3-(amino(3-chloro-4-(trifluoromethyl)phenyl)(cyclopropyl)methyl)-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | 分子名称: | 3-[(R)-amino[3-chloro-4-(trifluoromethyl)phenyl](cyclopropyl)methyl]-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETIC ACID, ... | | 著者 | Deng, X, Tomchick, D, Phillips, M. | | 登録日 | 2024-09-11 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKQ
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1153 (4,6-dicyclopropyl-3-(3-fluoro-4-(trifluoromethyl)benzyl)-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | 分子名称: | 4,6-dicyclopropyl-3-{[3-fluoro-4-(trifluoromethyl)phenyl]methyl}-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Tomchic, D, Phillips, M. | | 登録日 | 2024-09-09 | | 公開日 | 2025-01-01 | | 最終更新日 | 2025-01-22 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKO
 
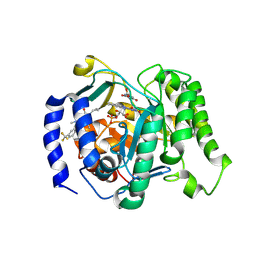 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1010 (6-cyclopropyl-2,4-dimethyl-3-(4-(trifluoromethyl)benzyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | 分子名称: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[4-(trifluoromethyl)phenyl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Tomchick, D, Phillips, M. | | 登録日 | 2024-09-09 | | 公開日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
3ZKX
 
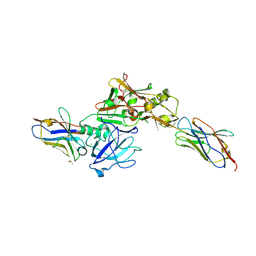 | | TERNARY BACE2 XAPERONE COMPLEX | | 分子名称: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | 登録日 | 2013-01-25 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZM6
 
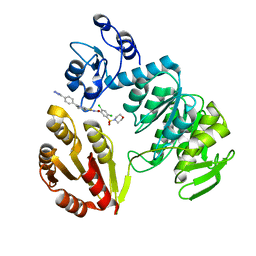 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | 分子名称: | N-(6-(4-(2h-tetrazol-5-yl)benzyl)-3-cyano-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-2,4-dichloro-5-(morpholinosulfonyl)benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | 著者 | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
9DFD
 
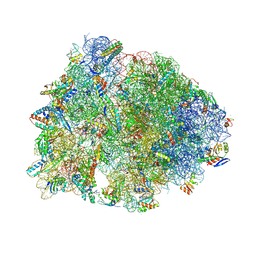 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with lasso peptide lariocidin B, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.60A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Aleksandrova, E.V, Travin, D.Y, Jangra, M, Kaur, M, Darwish, L, Koteva, K, Klepacki, D, Wang, W, Tiffany, M, Sokaribo, A, Coombes, B.K, Vazquez-Laslop, N, Wright, G.D, Mankin, A.S, Polikanov, Y.S. | | 登録日 | 2024-08-29 | | 公開日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome.
Res Sq, 2024
|
|
6WC6
 
 | |
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | 分子名称: | THIOCYANATE ION, hIAPP(15-25)WT | | 著者 | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | 登録日 | 2016-06-28 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | 主引用文献 | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
6WAQ
 
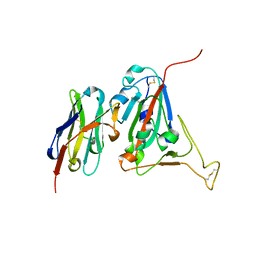 | |
7KNA
 
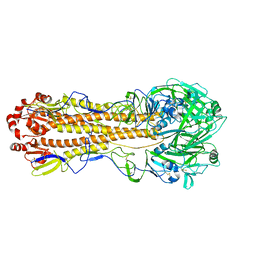 | | Localized reconstruction of the H1 A/Michigan/45/2015 ectodomain displayed at the surface of I53_dn5 nanoparticle | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,I53_dn5 | | 著者 | Acton, O.J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Quadrivalent influenza nanoparticle vaccines induce broad protection.
Nature, 592, 2021
|
|
6V4P
 
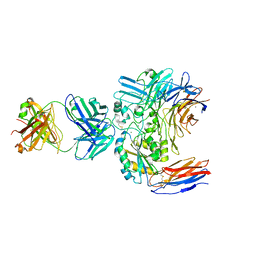 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | 分子名称: | Abciximab, heavy chain, light chain, ... | | 著者 | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | 登録日 | 2019-11-28 | | 公開日 | 2020-02-05 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
9EP8
 
 | | Crystal structure of ROCK2 in complex with a 8-(azaindolyl)-benzoazepinone inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 8-(azaindolyl)-benzoazepinone, Rho-associated protein kinase 2 | | 著者 | Pala, D, Clark, D, Pioselli, B, Accetta, A, Rancati, F. | | 登録日 | 2024-03-18 | | 公開日 | 2024-09-25 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Design and synthesis of novel 8-(azaindolyl)-benzoazepinones as potent and selective ROCK inhibitors.
Rsc Med Chem, 15, 2024
|
|
5MPP
 
 | | Structure of AaLS-wt | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | 登録日 | 2016-12-17 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5VWQ
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase | | 著者 | Mascarenhas, R, Lehrer, H, Liu, D, Ringe, D. | | 登録日 | 2017-05-22 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
3ZM5
 
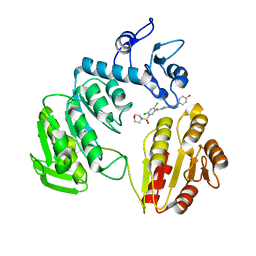 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | 分子名称: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | 著者 | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
9EWY
 
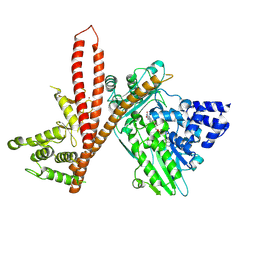 | | CryoEM structure of human MICAL1 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, ZINC ION, [F-actin]-monooxygenase MICAL1 | | 著者 | Schrofel, A, Pinkas, D, Novacek, J, Rozbesky, D. | | 登録日 | 2024-04-05 | | 公開日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of MICAL autoinhibition.
Nat Commun, 15, 2024
|
|
