5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Kovalevsky, A.Y, Gerlits, O.O. | | 登録日 | 2015-10-08 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5K
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | 著者 | Kovalevsky, A.Y, Das, A. | | 登録日 | 2015-10-08 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | 主引用文献 | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3TL9
 
 | | crystal structure of HIV protease model precursor/Saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3UHL
 
 | |
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-26 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3TKW
 
 | | Crystal structure of HIV protease model precursor/Darunavir complex | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
5T8H
 
 | |
1NAP
 
 | |
1TVX
 
 | |
3IBF
 
 | | Crystal structure of unliganded caspase-7 | | 分子名称: | Caspase-7 | | 著者 | Agniswamy, J. | | 登録日 | 2009-07-15 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformational similarity in the activation of caspase-3 and -7 revealed by the unliganded and inhibited structures of caspase-7.
Apoptosis, 14, 2009
|
|
3IBC
 
 | |
2G69
 
 | |
3I7E
 
 | | Co-crystal structure of HIV-1 protease bound to a mutant resistant inhibitor UIC-98038 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, HIV-1 protease | | 著者 | Hong, L, Tang, J, Ghosh, A. | | 登録日 | 2009-07-08 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
3EDR
 
 | |
3TWH
 
 | | Selenium Derivatized RNA/DNA Hybrid in complex with RNase H Catalytic Domain D132N Mutant | | 分子名称: | DNA (5'-D(*AP*TP*(SDG)P*TP*CP*(SDG))-3'), MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Rob, A, Gerlits, O, Jiang, J.S, Gan, J.H, Huang, Z. | | 登録日 | 2011-09-21 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Novel complex MAD phasing and RNase H structural insights using selenium oligonucleotides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PGA
 
 | | GLUTAMINASE-ASPARAGINASE FROM PSEUDOMONAS 7A | | 分子名称: | AMMONIUM ION, GLUTAMINASE-ASPARAGINASE, SULFATE ION | | 著者 | Jakob, C.G, Lewinski, K, Lacount, M.W, Roberts, J, Lebioda, L. | | 登録日 | 1997-01-14 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Ion binding induces closed conformation in Pseudomonas 7A glutaminase-asparaginase (PGA): crystal structure of the PGA-SO4(2-)-NH4+ complex at 1.7 A resolution.
Biochemistry, 36, 1997
|
|
4PHV
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HIV PROTEASE COMPLEX WITH L-700,417, AN INHIBITOR WITH PSEUDO C2 SYMMETRY | | 分子名称: | HIV-1 PROTEASE, N,N-BIS(2-HYDROXY-1-INDANYL)-2,6- DIPHENYLMETHYL-4-HYDROXY-1,7-HEPTANDIAMIDE | | 著者 | Bone, R. | | 登録日 | 1991-10-04 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-Ray Crystal Structure of the HIV Protease Complex with L-700,417, an Inhibitor with Pseudo C2 Symmetry
J.Am.Chem.Soc., 113, 1991
|
|
1QE6
 
 | | INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (L5C/H33C) | | 分子名称: | INTERLEUKIN-8 VARIANT, SULFATE ION | | 著者 | Gerber, N, Lowman, H, Artis, D.R, Eigenbrot, C. | | 登録日 | 1999-07-13 | | 公開日 | 2000-03-22 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Receptor-binding conformation of the "ELR" motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution.
Proteins, 38, 2000
|
|
2GZW
 
 | | Crystal structure of the E.coli CRP-cAMP complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | 著者 | Kumarevel, T.S, Tanaka, T, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-05-12 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of activated CRP protein from E coli
To be Published
|
|
6S1U
 
 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant | | 分子名称: | Gag-Pro-Pol polyprotein | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1V
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1CGP
 
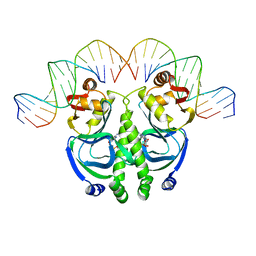 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G)-3'), ... | | 著者 | Schultz, S.C, Shields, G.C, Steitz, T.A. | | 登録日 | 1991-08-12 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees.
Science, 253, 1991
|
|
1I6X
 
 | | STRUCTURE OF A STAR MUTANT CRP-CAMP AT 2.2 A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | 著者 | White, M.A, Lee, J.C, Fox, R.O. | | 登録日 | 2001-03-06 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
1ICW
 
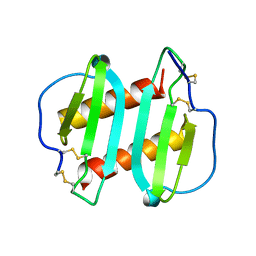 | | INTERLEUKIN-8, MUTANT WITH GLU 38 REPLACED BY CYS AND CYS 50 REPLACED BY ALA | | 分子名称: | INTERLEUKIN-8 | | 著者 | Eigenbrot, C, Lowman, H.B, Chee, L, Artis, D.R. | | 登録日 | 1996-09-18 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural change and receptor binding in a chemokine mutant with a rearranged disulfide: X-ray structure of E38C/C50AIL-8 at 2 A resolution.
Proteins, 27, 1997
|
|
