1U94
 
 | |
1U99
 
 | |
1U98
 
 | |
1XMS
 
 | | E. coli RecA in complex with MnAMP-PNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RecA protein | | Authors: | Bell, C.E, Xing, X. | | Deposit date: | 2004-10-04 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Escherichia coli RecA in Complex with MgADP and MnAMP-PNP(,).
Biochemistry, 43, 2004
|
|
1XMV
 
 | | E. coli RecA in complex with MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RecA protein | | Authors: | Bell, C.E, Xing, X. | | Deposit date: | 2004-10-04 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Escherichia coli RecA in Complex with MgADP and MnAMP-PNP(,).
Biochemistry, 43, 2004
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
2BC4
 
 | | Crystal structure of HLA-DM | | Descriptor: | CHLORIDE ION, HLA class II histocompatibility antigen, DM alpha chain, ... | | Authors: | Nicholson, M.J, Moradi, B, Seth, N.P, Xing, X, Cuny, G.D, Stein, R.L, Wucherpfennig, K.W. | | Deposit date: | 2005-10-18 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Small molecules that enhance the catalytic efficiency of HLA-DM.
J.Immunol., 176, 2006
|
|
7YBJ
 
 | | SARS-CoV-2 Mu variant spike(close state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBI
 
 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBL
 
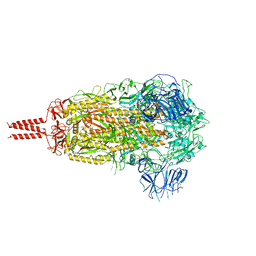 | | SARS-CoV-2 B.1.620 variant spike (close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBK
 
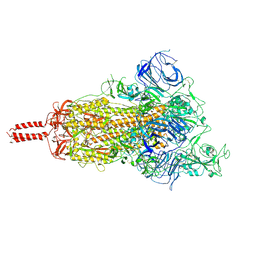 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBH
 
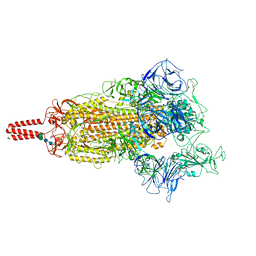 | | SARS-CoV-2 lambda variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBM
 
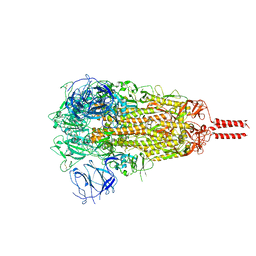 | | SARS-CoV-2 C.1.2 variant spike (Close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBN
 
 | | SARS-CoV-2 C.1.2 variant spike (Open state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
4BGG
 
 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP5
 
 | | Crystal structure of (S)-2-haloacid dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP7
 
 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8XOK
 
 | | Cryo-EM structure of human ABCC4 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, PALMITIC ACID | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
2QXV
 
 | | Structural basis of EZH2 recognition by EED | | Descriptor: | Embryonic ectoderm development, Enhancer of zeste homolog 2 | | Authors: | Han, Z. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of EZH2 recognition by EED
Structure, 15, 2007
|
|
2GAM
 
 | | X-ray crystal structure of murine leukocyte-type Core 2 b1,6-N-acetylglucosaminyltransferase (C2GnT-L) in complex with Galb1,3GalNAc | | Descriptor: | beta-1,6-N-acetylglucosaminyltransferase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystal Structure of Leukocyte Type Core 2 beta1,6-N-Acetylglucosaminyltransferase: Evidence for a covergence of metal ion independent glycosyltransferase mechanism.
J.Biol.Chem., 281, 2006
|
|
2GAK
 
 | |
7LH6
 
 | |
7LJJ
 
 | |
