8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of D-protein binder D-19437 and L-target L-Pep-1
To Be Published
|
|
4SLI
 
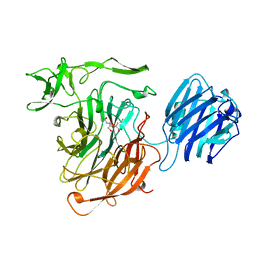 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2-PROPENYL-NEU5AC, AN INACTIVE SUBSTRATE ANALOGUE | | Descriptor: | 2-propenyl-N-acetyl-neuraminic acid, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-04 | | Release date: | 1999-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
2SLI
 
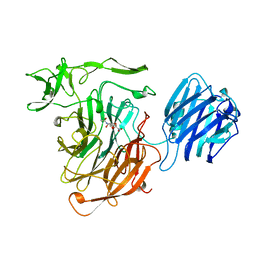 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC, THE REACTION PRODUCT | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
1DDM
 
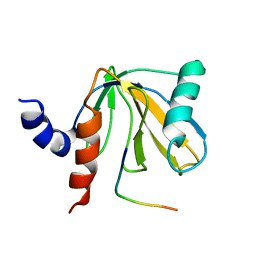 | | SOLUTION STRUCTURE OF THE NUMB PTB DOMAIN COMPLEXED TO A NAK PEPTIDE | | Descriptor: | NUMB ASSOCIATE KINASE, NUMB PROTEIN | | Authors: | Zwahlen, C, Li, S.C, Kay, L.E, Pawson, T, Forman-Kay, J.D. | | Deposit date: | 1999-11-11 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb.
EMBO J., 19, 2000
|
|
1G13
 
 | | HUMAN GM2 ACTIVATOR STRUCTURE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GANGLIOSIDE M2 ACTIVATOR PROTEIN | | Authors: | Wright, C.S, Li, S.C, Rastinejad, F. | | Deposit date: | 2000-10-10 | | Release date: | 2001-04-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human GM2-activator protein with a novel beta-cup topology.
J.Mol.Biol., 304, 2000
|
|
1SLL
 
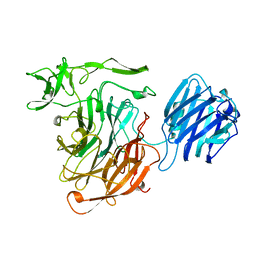 | | SIALIDASE L FROM LEECH MACROBDELLA DECORA | | Descriptor: | SIALIDASE L | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1997-10-14 | | Release date: | 1998-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
1SLI
 
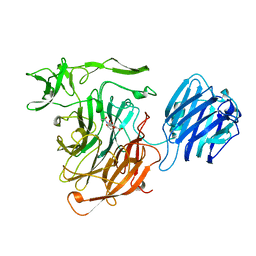 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1998-03-28 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
3SLI
 
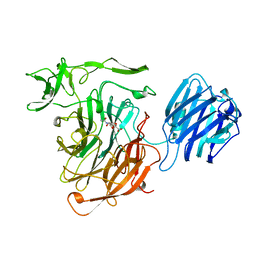 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC PREPARED BY SOAKING WITH 3'-SIALYLLACTOSE | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
2JMB
 
 | | Solution structure of the protein Atu4866 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu4866 | | Authors: | Ai, X, Semesi, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-11-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein Atu4866 from Agrobacterium tumefaciens
To be Published
|
|
2JPI
 
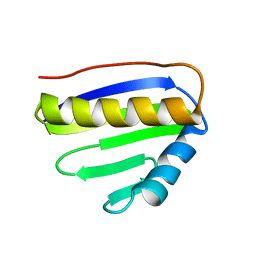 | | NMR structure of PA4090 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesi, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-05-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | chemical shift assignments of PA4090 from Pseudomonas aeruginosa
To be Published
|
|
2NMB
 
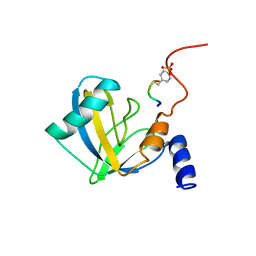 | | DNUMB PTB DOMAIN COMPLEXED WITH A PHOSPHOTYROSINE PEPTIDE, NMR, ENSEMBLE OF STRUCTURES. | | Descriptor: | PROTEIN (GPPY PEPTIDE), PROTEIN (NUMB PROTEIN) | | Authors: | Li, S.-C, Zwahlen, C, Vincent, S.J.F, McGlade, C.J, Pawson, T, Forman-Kay, J.D. | | Deposit date: | 1998-10-29 | | Release date: | 1998-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity.
Nat.Struct.Biol., 5, 1998
|
|
1KA7
 
 | | SAP/SH2D1A bound to peptide n-Y-c | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-Y-c | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KA6
 
 | | SAP/SH2D1A bound to peptide n-pY | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-pY | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1UPS
 
 | | GlcNAc[alpha]1-4Gal releasing endo-[beta]-galactosidase from Clostridium perfringens | | Descriptor: | CALCIUM ION, GLCNAC-ALPHA-1,4-GAL-RELEASING ENDO-BETA-GALACTOSIDASE | | Authors: | Tempel, W, Liu, Z.-J, Horanyi, P.S, Deng, L, Lee, D, Newton, M.G, Rose, J.P, Ashida, H, Li, S.-C, Li, Y.-T, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-25 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three-dimensional structure of GlcNAcalpha1-4Gal releasing endo-beta-galactosidase from Clostridium perfringens.
Proteins, 59, 2005
|
|
2AG4
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, Ganglioside GM2 activator, ISOPROPYL ALCOHOL, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG9
 
 | | Crystal Structure of the Y137S mutant of GM2-Activator Protein | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AGC
 
 | | Crystal Structure of mouse GM2- activator Protein | | Descriptor: | Ganglioside GM2 activator, LAURIC ACID, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AF9
 
 | | Crystal Structure analysis of GM2-Activator protein complexed with phosphatidylcholine | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, LAURIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG2
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with Phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
6E8M
 
 | |
6E8I
 
 | |
6E8H
 
 | | Legionella Longbeachae LeSH (Llo2327) | | Descriptor: | CHLORIDE ION, LeSH (Llo2327) | | Authors: | Kaneko, T, Li, S.S.C. | | Deposit date: | 2018-07-29 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and characterization of a large family of superbinding bacterial SH2 domains.
Nat Commun, 9, 2018
|
|
6E8K
 
 | |
2NNZ
 
 | | Solution structure of the hypothetical protein AF2241 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesib, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W.Y, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein AF2241 from Archaeoglobus fulgidus adopts a cyclophilin-like fold.
J.Biomol.Nmr, 38, 2007
|
|
5YPO
 
 | | Crystal structure of PSD-95 GK domain in complex with phospho-SAPAP peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, SAPAP | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhang, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
