5BRR
 
 | | Michaelis complex of tPA-S195A:PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Crystal Structure of the Michaelis Complex between Tissue-type Plasminogen Activator and Plasminogen Activators Inhibitor-1
J.Biol.Chem., 290, 2015
|
|
4XSK
 
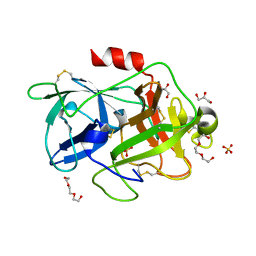 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
7TD5
 
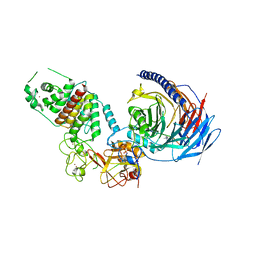 | | Structure of human PRC2-EZH1 containing phosphorylated SUZ12 | | Descriptor: | Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Gong, L, Jiao, L, Liu, X. | | Deposit date: | 2021-12-30 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site.
Nat Commun, 13, 2022
|
|
6W0B
 
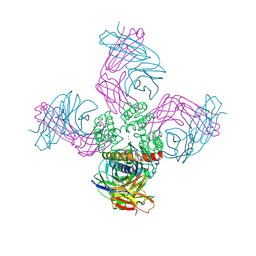 | | Open-gate KcsA soaked in 2 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.604 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0C
 
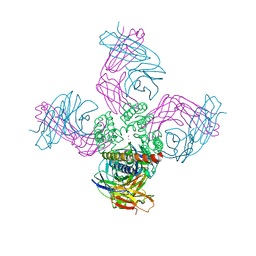 | | Open-gate KcsA soaked in 4 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0F
 
 | | Closed-gate KcsA soaked in 0mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0E
 
 | | Open-gate KcsA soaked in 10 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.512 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0H
 
 | | Closed-gate KcsA soaked in 5mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0G
 
 | | Closed-gate KcsA soaked in 1mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0D
 
 | | Open-gate KcsA soaked in 5 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.639 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0J
 
 | | Closed-gate KcsA incubated in BaCl2/NaCl | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0A
 
 | | Open-gate KcsA soaked in 1 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0I
 
 | | Closed-gate KcsA soaked in 10mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
4IZY
 
 | |
4G1W
 
 | |
5OFK
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J, Tu, T. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFL
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, SULFATE ION, ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFJ
 
 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
2XUM
 
 | | FACTOR INHIBITING HIF (FIH) Q239H MUTANT IN COMPLEX WITH ZN(II), NOG AND ASP-SUBSTRATE PEPTIDE (20-MER) | | Descriptor: | ASP-SUBSTRATE PEPTIDE 2, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor.
J. Biol. Chem., 286, 2011
|
|
6L94
 
 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
5XG4
 
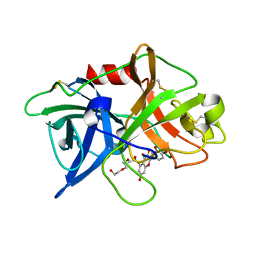 | | Crystal structure of uPA in complex with quercetin | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural mechanism of flavonoids in inhibiting serine proteases
Food Funct, 8, 2017
|
|
4HYS
 
 | |
4HYU
 
 | |
4NJ4
 
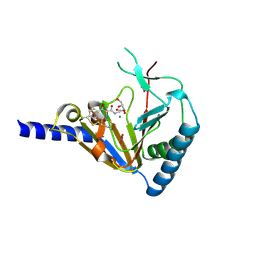 | | Crystal Structure of Human ALKBH5 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
