1UG7
 
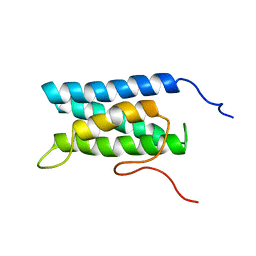 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | Descriptor: | 2610208M17Rik protein | | Authors: | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-13 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
5K19
 
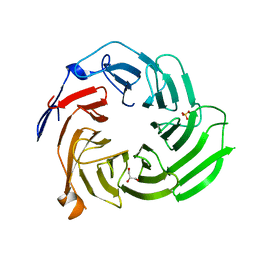 | |
5K1A
 
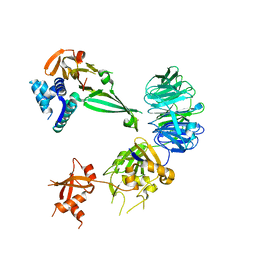 | |
4CWX
 
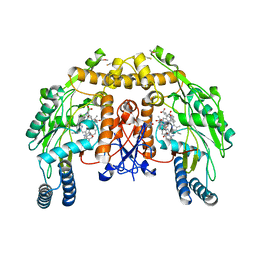 | | ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE COMPLEX1 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-{[5-(6-aminopyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CX7
 
 | | Structure of human iNOS heme domain in complex with (R)-6-(3-AMINO-2-(5-(2-(6-AMINO-4- METHYLPYRIDIN-2-YL)ETHYL)PYRIDIN-3-YL)PROPYL)-4- METHYLPYRIDIN-2-AMINE | | Descriptor: | (R)-6-(3-amino-2-(5-(2-(6-amino-4-methylpyridin-2-yl)ethyl)pyridin-3-yl)propyl)-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CX6
 
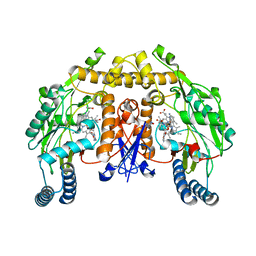 | | Structure of rat neuronal nitric oxide synthase H341L mutant heme domain in complex with 6-((((3S, 5R)-5-(((6-AMINO-4-METHYLPYRIDIN-2- YL)METHOXY)METHYL)PYRROLIDIN-3-YL)OXY)METHYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-((((3S, 5R)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CWW
 
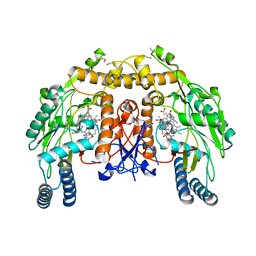 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 4-METHYL-6-(((3R,4R)-4-((5-(4-METHYLPYRIDIN-2-YL)PENTYL) OXY)PYRROLIDIN-3-YL)METHYL)PYRIDIN-2-AMINE | | Descriptor: | 4-methyl-6-{[(3R,4R)-4-{[5-(4-methylpyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CWY
 
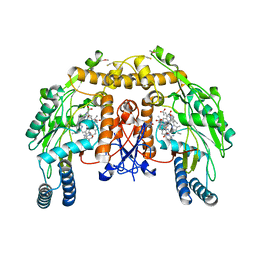 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6-(5-(((3R,4R)-4-((6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL) PYRROLIDIN-3-YL)OXY)PENTYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[5-({(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}oxy)pentyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CWV
 
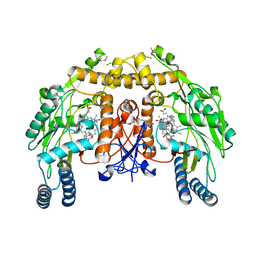 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 4-METHYL-6-(((3R,4R)-4-((5-(PYRIDIN-2-YL)PENTYL)OXY) PYRROLIDIN-3-YL)METHYL)PYRIDIN-2-AMINE | | Descriptor: | 4-methyl-6-{[(3R,4R)-4-{[5-(pyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CX1
 
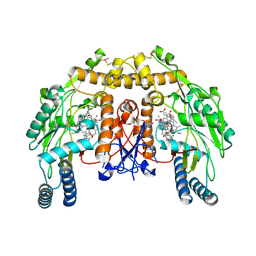 | | Structure of bovine endothelial nitric oxide synthase L111A mutant heme domain in complex with 4-METHYL-6-(((3R,4R)-4-((5-(4- METHYLPYRIDIN-2-YL)PENTYL)OXY)PYRROLIDIN-3-YL)METHYL)PYRIDIN-2-AMINE | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, 4-methyl-6-{[(3R,4R)-4-{[5-(4-methylpyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
4CX2
 
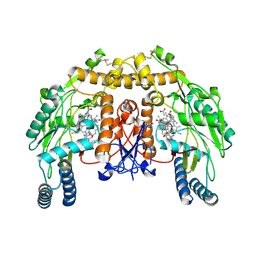 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6-(5-(((3R,4R)-4-((6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL) PYRROLIDIN-3-YL)OXY)PENTYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, 6-[5-({(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}oxy)pentyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
3D1D
 
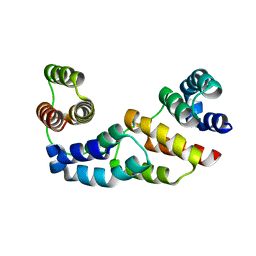 | | Hexagonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
6EDQ
 
 | | Crystal Structure of the Light-Gated Anion Channelrhodopsin GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin 1, GLYCEROL | | Authors: | Li, H, Huang, C.Y, Wang, M, Zheng, L, Spudich, J.L. | | Deposit date: | 2018-08-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a natural light-gated anion channelrhodopsin.
Elife, 8, 2019
|
|
3D1B
 
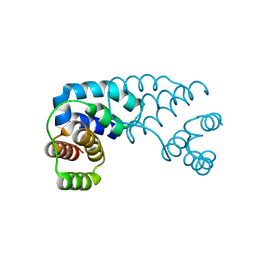 | | Tetragonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
4LNQ
 
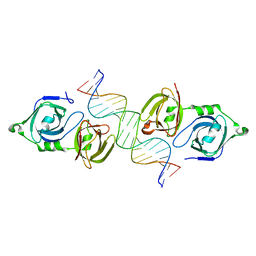 | |
3S8M
 
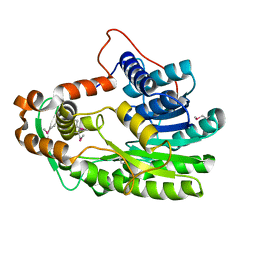 | | The Crystal Structure of FabV | | Descriptor: | Enoyl-ACP Reductase | | Authors: | Li, H, Zhang, X.L, Bi, L.J, He, J, Jiang, T. | | Deposit date: | 2011-05-29 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determination of the Crystal Structure and Active Residues of FabV, the Enoyl-ACP Reductase from Xanthomonas oryzae.
Plos One, 6, 2011
|
|
1UEZ
 
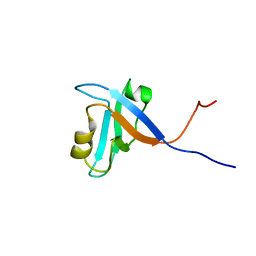 | | Solution structure of the first PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 Protein | | Authors: | Li, H, Kigawa, T, Muto, Y, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first PDZ domain of human KIAA1526 protein
To be Published
|
|
1UEM
 
 | | Solution Structure of the First Fibronectin Type III domain of human KIAA1568 Protein | | Descriptor: | KIAA1568 Protein | | Authors: | Li, H, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First Fibronectin Type III domain of human KIAA1568 Protein
To be Published
|
|
9NSE
 
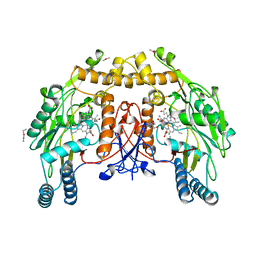 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, ETHYL-ISOSELENOUREA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2000-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
4D1O
 
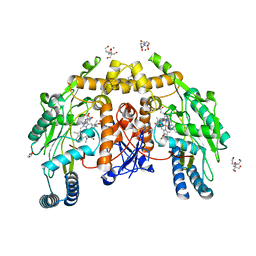 | | Structure of human endothelial nitric oxide synthase heme domain with L-Arg bound | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structures of Human Constitutive Nitric Oxide Synthases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D1P
 
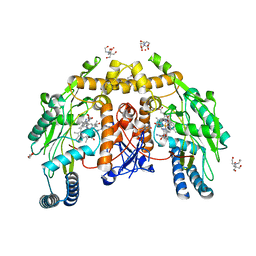 | | Structure of human endothelial nitric oxide synthase heme domain IN COMPLEX WITH 6-((((3S, 5R)-5-(((6-AMINO-4-METHYLPYRIDIN-2-YL)METHOXY) METHYL)PYRROLIDIN-3-YL)OXY) METHYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-((((3S, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Structures of Human Constitutive Nitric Oxide Synthases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D1N
 
 | | Structure of human nNOS heme domain with L-Arg bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of Human Constitutive Nitric Oxide Synthases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XPY
 
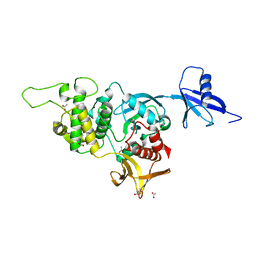 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
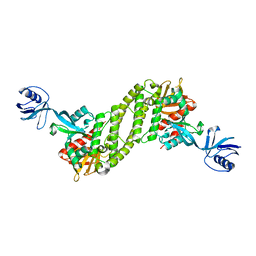 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
