5A0N
 
 | | N-terminal thioester domain of protein F2 like fibronectin-binding protein from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PROTEIN F2 LIKE FIBRONECTIN-BINDING PROTEIN | | Authors: | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
5ACB
 
 | | Crystal Structure of the Human Cdk12-Cyclink Complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Mackenzie, A, Goubin, S, Strain-Damerell, C, Mahajan, P, Tallant, C, Chalk, R, Wiggers, H, Kopec, J, Fitzpatrick, F, Burgess-Brown, N, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-15 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Targeting of Remote Cysteine Residues to Develop Cdk12 and Cdk13 Inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
6VKO
 
 | |
5J32
 
 | | Isopropylmalate dehydrogenase in complex with isopropylmalate | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase 2, chloroplastic, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
6V2E
 
 | |
6VCJ
 
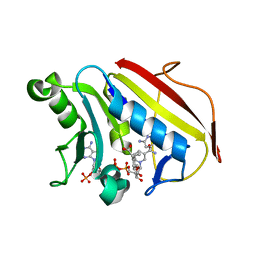 | | Crystal structure of hsDHFR in complex with NADP+, DAP, and R-naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Pedersen, L.C, London, R.E, Gabel, S.A, Krahn, J.M, DeRose, E.F. | | Deposit date: | 2019-12-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Structural Basis for Nonsteroidal Anti-Inflammatory Drug Inhibition of Human Dihydrofolate Reductase.
J.Med.Chem., 63, 2020
|
|
5JDV
 
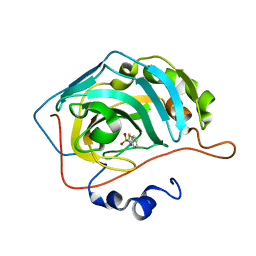 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6VE6
 
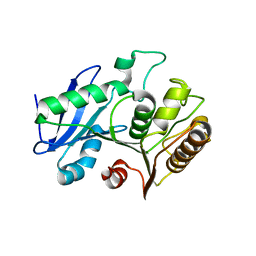 | | A structural characterization of poly(aspartic acid) hydrolase-1 from Sphingomonas sp. KT-1. | | Descriptor: | Poly(Aspartic acid) hydrolase-1 | | Authors: | Bolay, A.L, Salvo, H, Brambley, C.A, Yared, T.J, Miller, J.M, Wallen, J.R, Weiland, M.H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Structural Characterization of Sphingomonas sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
Acs Sustain Chem Eng, 2020
|
|
4YUO
 
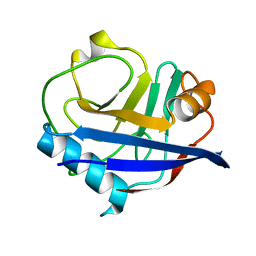 | | High-resolution multiconformer synchrotron model of CypA at 273 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
5JEH
 
 | | Human carbonic anhydrase II (L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6UP1
 
 | |
5J33
 
 | | Isopropylmalate dehydrogenase in complex with NAD+ | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
4YUJ
 
 | | Multiconformer synchrotron model of CypA at 240 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YVM
 
 | |
4Y6W
 
 | |
1GGI
 
 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6VL2
 
 | | Stigmurin | | Descriptor: | Stigmurin | | Authors: | Rodrigues, S.C.S, Resende, J.M, Araujo, R.M, Pedrosa, M.F.F. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-13 | | Last modified: | 2021-01-20 | | Method: | SOLUTION NMR | | Cite: | NMR three-dimensional structure of the cationic peptide Stigmurin from Tityus stigmurus scorpion venom: In vitro antioxidant and in vivo antibacterial and healing activity.
Peptides, 137, 2021
|
|
4YHR
 
 | | Crystal Structure of Yeast Proliferating Cell Nuclear Antigen | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Litman, J.M, Nguyen, V.Q, Kondratick, C.M, Powers, K.T, Schnieders, M.J, Washington, M.T. | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9502 Å) | | Cite: | Dead-End Elimination with a Polarizable Force Field Repacks PCNA Models from Low-Resolution X-ray Diffraction into Atomic Resolution Structures
To be published
|
|
5JG3
 
 | | Human carbonic anhydrase II (121T/N67Q) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JGS
 
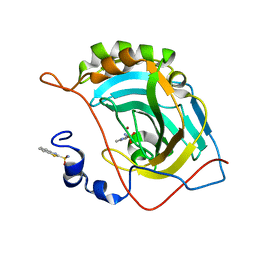 | | Human carbonic anhydrase II (F131Y/L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5AIK
 
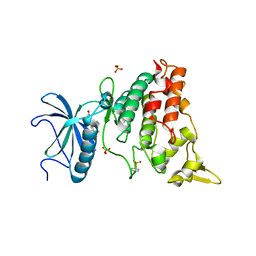 | | Human DYRK1A in complex with LDN-211898 | | Descriptor: | 4-(7-METHOXY-1-(TRIFLUOROMETHYL)-9H-PYRIDO[3,4-B]INDOL-9-yl)butan-1-amine, DYRK1A DUAL-SPECIFICITY TYROSINE-PHOSPHORYLATION REGULATED KINASE 1A, PHOSPHATE ION | | Authors: | Elkins, J.M, Soundararajan, M, Muniz, J.R.C, Cuny, G, Higgins, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dyrk1A with Ldn-211898
To be published
|
|
5A3G
 
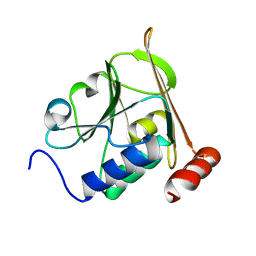 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6TAE
 
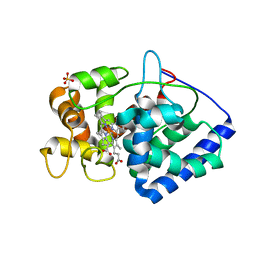 | | Neutron structure of ferric ascorbate peroxidase | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A1K
 
 | |
6TCQ
 
 | | Crystal structure of the omalizumab Fab Ser81Arg and Gln83Arg light chain mutant | | Descriptor: | GLYCEROL, Omalizumab Fab Ser81Arg and Gln83Arg light chain mutant | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
