4PD7
 
 | | Structure of vcCNT bound to zebularine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD8
 
 | | Structure of vcCNT-7C8C bound to pyrrolo-cytidine | | Descriptor: | 6-methyl-3-(beta-D-ribofuranosyl)-3,7-dihydro-2H-pyrrolo[2,3-d]pyrimidin-2-one, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD6
 
 | | Crystal structure of vcCNT-7C8C bound to uridine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD5
 
 | | Crystal structure of vcCNT-7C8C bound to gemcitabine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, GEMCITABINE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PDA
 
 | | Structure of vcCNT-7C8C bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD9
 
 | | Structure of vcCNT-7C8C bound to adenosine | | Descriptor: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PB1
 
 | | Structure of vcCNT-7C8C bound to ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
6TW3
 
 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | Descriptor: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | Descriptor: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Marsh, M.E, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8C0P
 
 | |
8C0S
 
 | |
8CF3
 
 | |
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D, ZINC ION | | Authors: | Ha, N.C, Kim, J.S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
5C21
 
 | | Crystal structure of native HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D | | Authors: | Ha, N.C, Kim, J.S, Yoon, B.Y. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
8C54
 
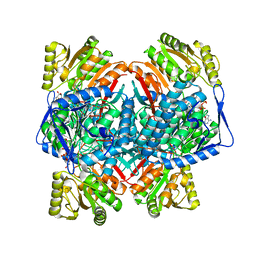 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
5KUC
 
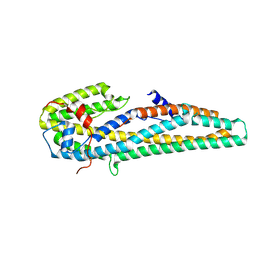 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
2JTN
 
 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
8DA2
 
 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
5XJG
 
 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DUA
 
 | | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, Envelope glycoprotein gp41, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-27 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of prefusion SIV envelope trimer.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DVD
 
 | |
