3R6L
 
 | | Caspase-2 T380A bound with Ac-VDVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)VDVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R5J
 
 | |
3R7B
 
 | | Caspase-2 bound to one copy of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7N
 
 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
4I9O
 
 | | Crystal Structure of GACKIX L664C Tethered to 1-10 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one, CREB-binding protein | | Authors: | Wang, N, Meagher, J.L, Stuckey, J.A, Mapp, A.K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ordering a dynamic protein via a small-molecule stabilizer.
J.Am.Chem.Soc., 135, 2013
|
|
6XKA
 
 | |
8SMK
 
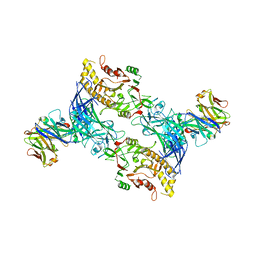 | | hPAD4 bound to Activating Fab hA362 | | Descriptor: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SML
 
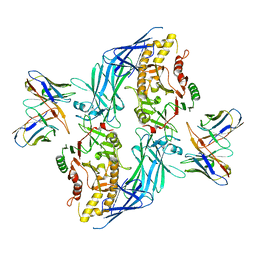 | | hPAD4 bound to inhibitory Fab hI365 | | Descriptor: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
6VPG
 
 | |
6VPM
 
 | |
6VPI
 
 | |
6VPJ
 
 | |
6VPH
 
 | |
6VPL
 
 | |
6VO4
 
 | | Crystal Structure Analysis of BFL1 | | Descriptor: | Bcl-2-related protein A1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Identification of a Covalent Molecular Inhibitor of Anti-apoptotic BFL-1 by Disulfide Tethering.
Cell Chem Biol, 27, 2020
|
|
2ST1
 
 | |
1ZDA
 
 | |
1ZDD
 
 | |
1ZDC
 
 | |
1ZDB
 
 | |
1ANP
 
 | |
1VPF
 
 | | STRUCTURE OF HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR | | Authors: | Muller, Y.A, De Vos, A.M. | | Deposit date: | 1997-04-08 | | Release date: | 1998-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1CA0
 
 | | BOVINE CHYMOTRYPSIN COMPLEXED TO APPI | | Descriptor: | BOVINE CHYMOTRYPSIN, PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | Authors: | Scheidig, A.J, Kossiakoff, A.A. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
1CBW
 
 | | BOVINE CHYMOTRYPSIN COMPLEXED TO BPTI | | Descriptor: | BOVINE CHYMOTRYPSIN, BPTI, SULFATE ION | | Authors: | Hynes, T.R, Scheidig, A.J, Kossiakoff, A.A. | | Deposit date: | 1996-12-22 | | Release date: | 1997-07-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
1FLT
 
 | | VEGF IN COMPLEX WITH DOMAIN 2 OF THE FLT-1 RECEPTOR | | Descriptor: | FMS-LIKE TYROSINE KINASE 1, VASCULAR ENDOTHELIAL GROWTH FACTOR | | Authors: | Wiesmann, C, De Vos, A.M. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor.
Cell(Cambridge,Mass.), 91, 1997
|
|
