5TJZ
 
 | | Structure of 4-Hydroxytetrahydrodipicolinate Reductase from Mycobacterium tuberculosis with NADPH and 2,6 Pyridine Dicarboxylic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, 4-hydroxy-tetrahydrodipicolinate reductase, ... | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
3C2S
 
 | | Structural Characterization of a Human Fc Fragment Engineered for Lack of Effector Functions | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2008-01-25 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of a human Fc fragment engineered for lack of effector functions.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5TEM
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Vibrio vulnificus with 2,6 Pyridine Dicarboxylic and NADH | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Mank, N, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
5TJY
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Mycobacterium tuberculosis with 2,6 Pyridine Dicarboxylic Acid and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-hydroxy-tetrahydrodipicolinate reductase, ... | | Authors: | Mank, N, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-10-05 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6UPJ
 
 | | HIV-2 PROTEASE/U99294 COMPLEX | | Descriptor: | 6,7,8,9-TETRAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOHEPTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
5UGV
 
 | | DapB from Mycobacterium tuberculosis | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Pote, S.P, Mank, N, Chruszcz, M. | | Deposit date: | 2017-01-10 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
5TEJ
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Vibrio vulnificus with 2,5 Furan Dicarboxylic and NADH | | Descriptor: | 2,5 Furan Dicarboxylic Acid, 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mank, N, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
5US6
 
 | | Structure of Dihydrodipicolinate Reductase from Vibrio vulnificus Bound to NADH and 2,6 Pyridine Dicarboxylic Acid with Intact Polyhistidine Tag | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Mank, N.M, Arnette, A.K, Chruszcz, M. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
1MOE
 
 | | The three-dimensional structure of an engineered scFv T84.66 dimer or diabody in VL to VH linkage. | | Descriptor: | SULFATE ION, anti-CEA mAb T84.66 | | Authors: | Carmichael, J.A, Power, B.E, Garrett, T.P.J, Yazaki, P.J, Shively, J.E, Raubischek, A.A, Wu, A.M, Hudson, P.J. | | Deposit date: | 2002-09-09 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of an Anti-CEA scFv Diabody Assembled from T84.66 scFvs in VL-to-VH Orientation: Implications for Diabody Flexibility
J.Mol.Biol., 326, 2003
|
|
3FSR
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP-dependent alcohol dehydrogenase, ... | | Authors: | Frolow, F, Greenblatt, H, Goihberg, E, Burstein, Y. | | Deposit date: | 2009-01-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
5HEZ
 
 | | JAK2 kinase (JH1 domain) mutant P1057A in complex with TG101209 | | Descriptor: | CHLORIDE ION, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide, Tyrosine-protein kinase JAK2, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity.
Sci Transl Med, 8, 2016
|
|
7VBM
 
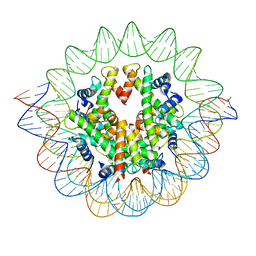 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
7DBH
 
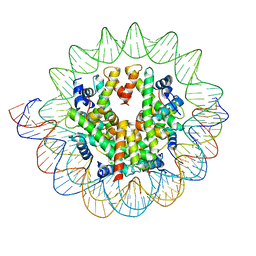 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
5VG2
 
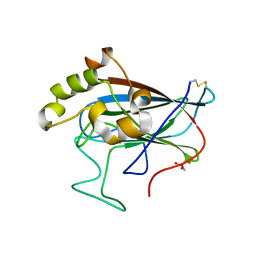 | | Intradiol ring-cleavage Dioxygenase from Tetranychus urticae | | Descriptor: | CACODYLATE ION, FE (III) ION, Intradiol ring-cleavage Dioxygenase | | Authors: | Schlachter, C, Klapper, V, Chruszcz, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch.
Insect Biochem.Mol.Biol., 107, 2019
|
|
1P91
 
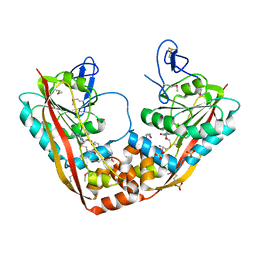 | | Crystal Structure of RlmA(I) enzyme: 23S rRNA n1-G745 methyltransferase (NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER19) | | Descriptor: | Ribosomal RNA large subunit methyltransferase A, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Das, K, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5UPJ
 
 | | HIV-2 PROTEASE/U99283 COMPLEX | | Descriptor: | 5,6,7,8,9,10-HEXAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOOCTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
7UPJ
 
 | | HIV-1 PROTEASE/U101935 COMPLEX | | Descriptor: | HIV-1 PROTEASE, N-(3-CYCLOPROPYL(5,6,7,8,9,10-HEXAHYDRO-2-OXO-2H-CYCLOOCTA[B]PYRAN-3-YL)METHYL)PHENYLBENZENSULFONAMIDE | | Authors: | Watenpaugh, K.D, Janakiraman, M.N. | | Deposit date: | 1996-12-05 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of nonpeptidic HIV protease inhibitors: the sulfonamide-substituted cyclooctylpyramones.
J.Med.Chem., 40, 1997
|
|
5YJ9
 
 | | Crystal structure of Tribolium castaneum PINK1 kinase domain in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PINK1, ... | | Authors: | Okatsu, K, Sato, Y, Fukai, S. | | Deposit date: | 2017-10-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into ubiquitin phosphorylation by PINK1.
Sci Rep, 8, 2018
|
|
2NVB
 
 | | Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases (ADHs) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Goihberg, E, Tel-Or, S, Peretz, M, Frolow, F, Dym, O, Burstein, Y, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution
Proteins, 72, 2008
|
|
2M6Q
 
 | | Refined Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Strucutral Genomics Consortium Target ZR18 | | Descriptor: | SAV1430 | | Authors: | Baran, M.C, Aramini, J.M, Huang, Y.J, Xiao, R, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
1YDO
 
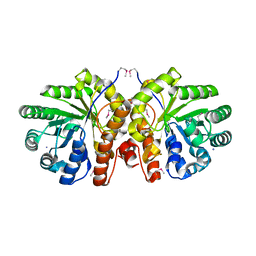 | | Crystal Structure of the Bacillis subtilis HMG-CoA Lyase, Northeast Structural Genomics Target SR181. | | Descriptor: | HMG-CoA Lyase, IODIDE ION | | Authors: | Forouhar, F, Hussain, M, Edstrom, W, Vorobiev, S.M, Xiao, R, Ciano, M, Shih, L, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J.Biol.Chem., 281, 2006
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
1YWL
 
 | |
3ZPF
 
 | |
3ZPE
 
 | |
