7KFV
 
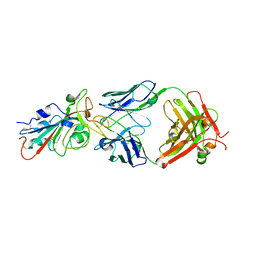 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFY
 
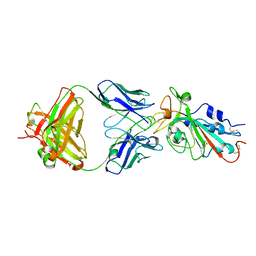 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFW
 
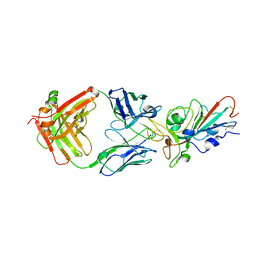 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFX
 
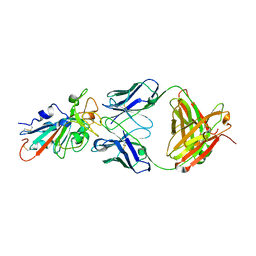 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
4HR9
 
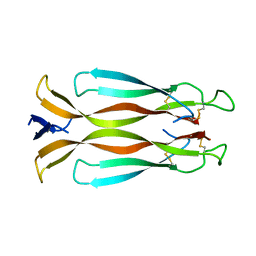 | | Human interleukin 17A | | Descriptor: | Interleukin-17A | | Authors: | Liu, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-05-22 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
4HSA
 
 | | Structure of interleukin 17a in complex with il17ra receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Liu, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
6JK2
 
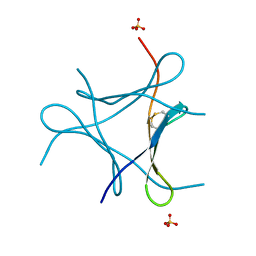 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6JK3
 
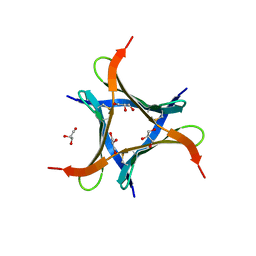 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
7E0W
 
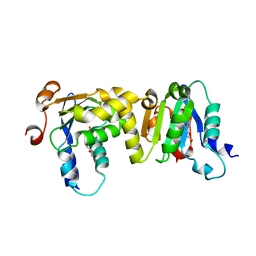 | | Crystal Structure of BCH domain from S. pombe | | Descriptor: | Putative Rho GTPase-activating protein C1565.02c, TETRAETHYLENE GLYCOL | | Authors: | Chichili, V.P.R, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel intertwined anti-parallel dimeric structure of scaffold BCH domain regulates RhoA and RhoGAP functions
Proc.Natl.Acad.Sci.USA, 2021
|
|
5IKF
 
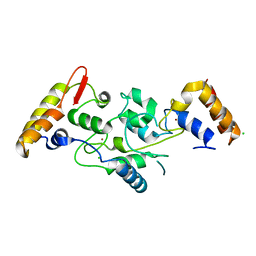 | |
5Y63
 
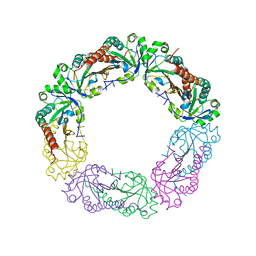 | | Crystal structure of Enterococcus faecalis AhpC | | Descriptor: | Alkyl hydroperoxide reductase, C subunit | | Authors: | Pan, A, Balakrishna, A.M, Grueber, G. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Atomic structure and enzymatic insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit C
Free Radic. Biol. Med., 115, 2017
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
5IKJ
 
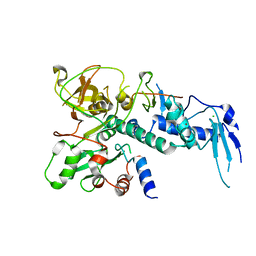 | | Structure of Clr2 bound to the Clr1 C-terminus | | Descriptor: | CHLORIDE ION, Cryptic loci regulator 2, Cryptic loci regulator protein 1, ... | | Authors: | Pfister, Y, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
5IKK
 
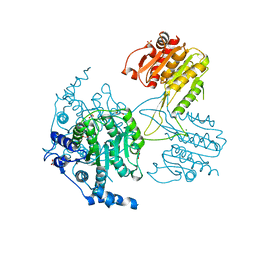 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
5VDR
 
 | |
5VDP
 
 | |
5VDO
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 2',2'-cGAMP | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.218 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
5VDU
 
 | |
5VDV
 
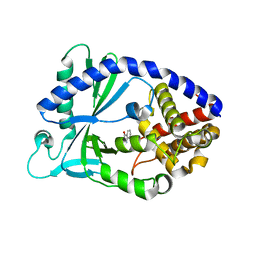 | |
5VDT
 
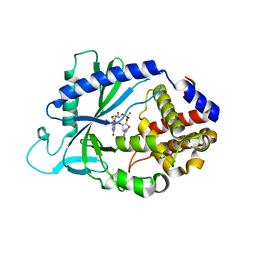 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
4WZ6
 
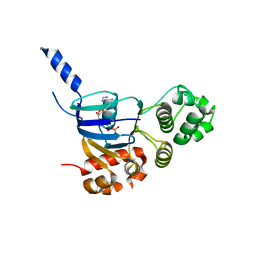 | | Human CFTR aa389-678 (NBD1), deltaF508 with three solubilizing mutations, bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Byrnes, L.J, Hall, J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy.
Protein Sci., 25, 2016
|
|
5VDS
 
 | |
5VDW
 
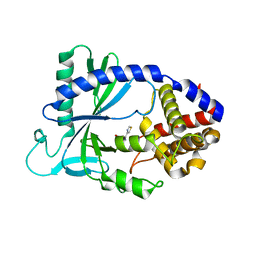 | | Human cyclic GMP-AMP synthase (cGAS) in complex with Compound F1 | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, [2-(1,3-thiazol-4-yl)-1H-benzimidazol-1-yl]acetic acid | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
5VDQ
 
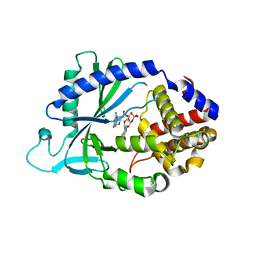 | |
5A9Y
 
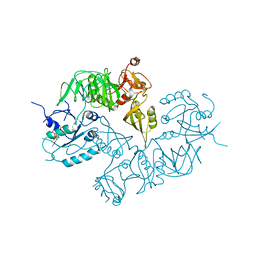 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
