6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6VBX
 
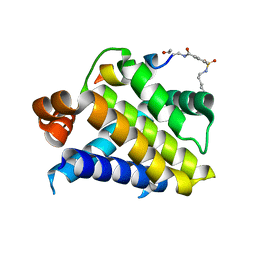 | | Crystal structure of Mcl-1 in complex with 138E12 peptide, Lys-covalent antagonist | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic peptide | | Authors: | Pellecchia, M, Perry, J.J, Kenjic, N, Assar, Z. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
J.Med.Chem., 64, 2021
|
|
6UP0
 
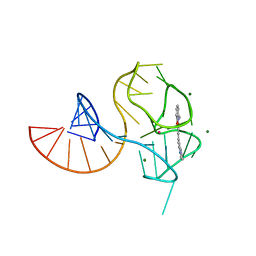 | |
6VIF
 
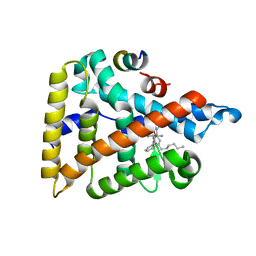 | | Human LRH-1 ligand-binding domain bound to agonist cpd 15 and fragment of coregulator TIF-2 | | Descriptor: | N-[(8beta,11alpha,12alpha)-8-{[methyl(phenyl)amino]methyl}-1,6:7,14-dicycloprosta-1(6),2,4,7(14)-tetraen-11-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of a new class of liver receptor homolog-1 (LRH-1) agonists by photoredox conjugate addition.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7PPM
 
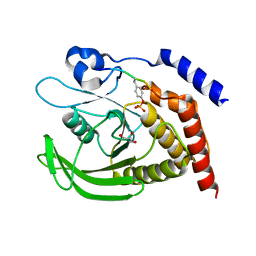 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPN
 
 | | SHP2 catalytic domain in complex with CD28 (183-198) phosphopeptide (pTyr-191, p-Thr-195) | | Descriptor: | GLYCEROL, T-cell-specific surface glycoprotein CD28, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPL
 
 | | SHP2 catalytic domain in complex with IRS1 (625-639) phosphopeptide (pTyr-632, pSer-636) | | Descriptor: | ETHANOL, GLYCEROL, Insulin receptor substrate 1, ... | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
6OPJ
 
 | | Menin in complex with peptide inhibitor 25 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, Peptide inhibitor 25, ... | | Authors: | Linhares, B.M, Fortuna, P, Cierpicki, T, Grembecka, J, Berlicki, L. | | Deposit date: | 2019-04-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5006572 Å) | | Cite: | Covalent and noncovalent constraints yield a figure eight-like conformation of a peptide inhibiting the menin-MLL interaction.
Eur.J.Med.Chem., 207, 2020
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
6PHJ
 
 | |
6PHO
 
 | |
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
6PHK
 
 | |
6PHP
 
 | |
6PQ7
 
 | |
6PHN
 
 | |
6PHI
 
 | |
6PHM
 
 | |
6PHL
 
 | |
6PHQ
 
 | |
6P7M
 
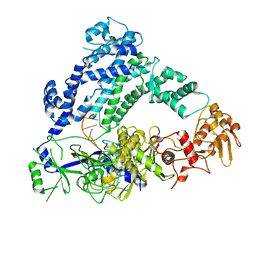 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
7QYH
 
 | |
7R7M
 
 | |
