8CRC
 
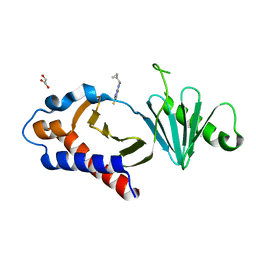 | | Structure of human Plk1 PBD in complex with Allopole-A | | Descriptor: | 7-chloro-4-(cyclopropylmethyl)-1-thioxo-2,4-dihydrothieno[2,3-e][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Kirsch, K, Park, J.E, Lee, K.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specific inhibition of an anticancer target, polo-like kinase 1, by allosterically dismantling its mechanism of substrate recognition.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | Descriptor: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
1HRM
 
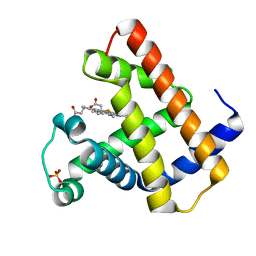 | |
5NNV
 
 | |
5NMO
 
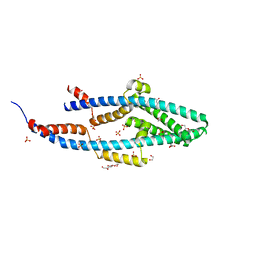 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
8GUW
 
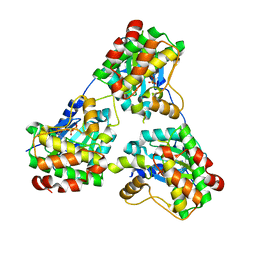 | |
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8IVU
 
 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
2L5N
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Barb, A.W, Lee, H, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C.R, Haleema, J, Acton, T.B, Xiao, R.R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
4RSI
 
 | | Yeast Smc2-Smc4 hinge domain with extended coiled coils | | Descriptor: | PHOSPHATE ION, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Soh, Y.M, Shin, H.C, Oh, B.H. | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for SMC Rod Formation and Its Dissolution upon DNA Binding.
Mol.Cell, 57, 2015
|
|
4RSJ
 
 | |
2XA9
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horikoshii in complex with UDPG | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2X6Q
 
 | | Crystal structure of trehalose synthase TreT from P.horikoshi | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA1
 
 | | Crystal structure of trehalose synthase TreT from P.horikoshii (Seleno derivative) | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
5B26
 
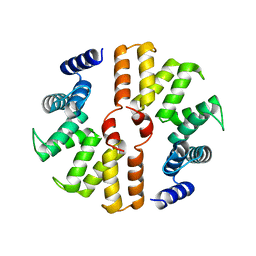 | | Crystal structure of mouse SEL1L | | Descriptor: | Protein sel-1 homolog 1 | | Authors: | Jeong, H, Lee, C. | | Deposit date: | 2016-01-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway
Sci Rep, 6, 2016
|
|
5GPG
 
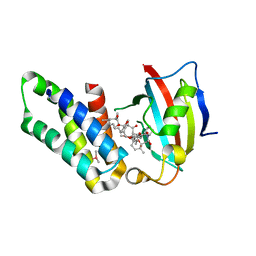 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
2QMQ
 
 | |
2SNW
 
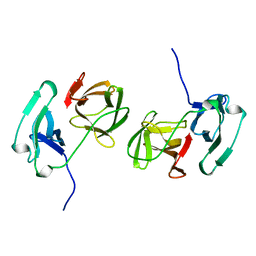 | | SINDBIS VIRUS CAPSID PROTEIN, TYPE3 CRYSTAL FORM | | Descriptor: | COAT PROTEIN C | | Authors: | Choi, H.-K, Lee, S, Zhang, Y.-P, Mckinney, B.R, Wengler, G, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 1998-02-17 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis.
J.Mol.Biol., 262, 1996
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
6PBA
 
 | | Structure of ClpC1-NTD | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1 | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBQ
 
 | | Structure of ClpC1-NTD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp protease ATP-binding subunit ClpC1, PHOSPHATE ION | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBS
 
 | | Structure of ClpC1-NTD in complex with Ecumicin | | Descriptor: | ACETATE ION, ATP-dependent Clp protease ATP-binding subunit ClpC1, ecumicin | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5YX3
 
 | |
5YX4
 
 | | Isoliquiritigenin-complexed Chalcone isomerase (S189A) from the Antarctic vascular plant Deschampsia Antarctica (DaCHI1) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Chalcone-flavonone isomerase family protein | | Authors: | Lee, C.W, Park, S, Lee, J.H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzymatic properties of chalcone isomerase from the Antarctic vascular plant Deschampsia antarctica Desv.
PLoS ONE, 13, 2018
|
|
