1YEE
 
 | |
1YEC
 
 | |
4Q21
 
 | |
1YTK
 
 | |
1YTD
 
 | |
1YT5
 
 | |
1YTE
 
 | |
1S7O
 
 | | Crystal structure of putative DNA binding protein SP_1288 from Streptococcus pygenes | | Descriptor: | Hypothetical UPF0122 protein SPy1201/SpyM3_0842/SPs1042/spyM18_1152 | | Authors: | Oganesyan, V, Pufan, R, DeGiovanni, A, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
3C4F
 
 | |
5DH4
 
 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3C4C
 
 | | B-Raf Kinase in Complex with PLX4720 | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide | | Authors: | Zhang, K.Y.J, Wang, W. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C4E
 
 | | Pim-1 Kinase Domain in Complex with 3-aminophenyl-7-azaindole | | Descriptor: | IMIDAZOLE, N-phenyl-1H-pyrrolo[2,3-b]pyridin-3-amine, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Zhang, K.Y.J, Wang, W. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5GGU
 
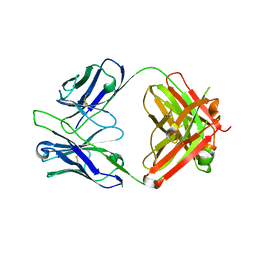 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGS
 
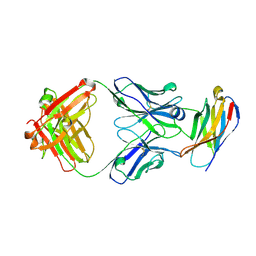 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGV
 
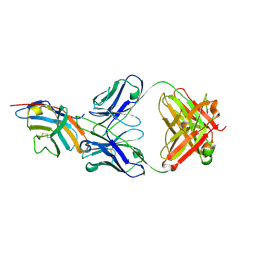 | | CTLA-4 in complex with tremelimumab Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
1SU0
 
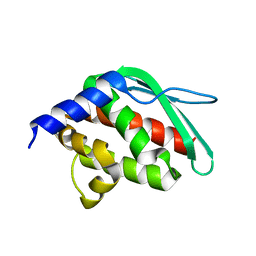 | | Crystal structure of a hypothetical protein at 2.3 A resolution | | Descriptor: | NifU like protein IscU, ZINC ION | | Authors: | Liu, J, Oganesyan, N, Shin, D.-H, Jancarik, J, Pufan, R, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form.
Proteins, 59, 2005
|
|
1T8B
 
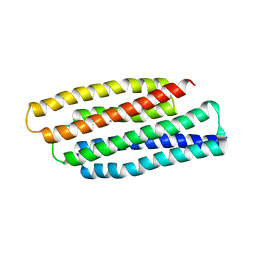 | | Crystal structure of refolded PHOU-like protein (gi 2983430) from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
1QU7
 
 | |
1RQ0
 
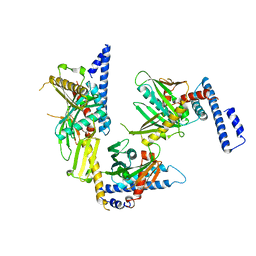 | | Crystal structure of peptide releasing factor 1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Shin, D.H, Brandsen, J, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome.
J.Mol.Biol., 341, 2004
|
|
1T72
 
 | | Crystal structure of phosphate transport system protein phoU from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
2EXM
 
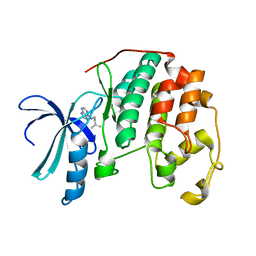 | | Human CDK2 in complex with isopentenyladenine | | Descriptor: | Cell division protein kinase 2, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE | | Authors: | Schulze-Gahmen, U. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple modes of ligand recognition: crystal structures of cyclin-dependent protein kinase 2 in complex with ATP and two inhibitors, olomoucine and isopentenyladenine.
Proteins, 22, 1995
|
|
255D
 
 | |
2RDB
 
 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase I100W Mutant | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Murray, L.J, Garcia-Serres, R, McCormick, M.S, Davydov, R, Naik, S, Hoffman, B.M, Huynh, B.H, Lippard, S.J. | | Deposit date: | 2007-09-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dioxygen activation at non-heme diiron centers: oxidation of a proximal residue in the I100W variant of toluene/o-xylene monooxygenase hydroxylase.
Biochemistry, 46, 2007
|
|
5I0B
 
 | | Structure of PAK4 | | Descriptor: | 6-bromo-2-[1-methyl-3-(propan-2-yl)-1H-pyrazol-4-yl]-1H-imidazo[4,5-b]pyridine, Serine/threonine-protein kinase PAK 4 | | Authors: | Park, S.Y. | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The discovery and the structural basis of an imidazo[4,5-b]pyridine-based p21-activated kinase 4 inhibitor
Bioorg. Med. Chem. Lett., 26, 2016
|
|
